Contribution of Gag and Protease to HIV-1 Phenotypic Drug Resistance in Pediatric Patients Failing Protease Inhibitor-Based Therapy
- PMID: 26833162
- PMCID: PMC4808165
- DOI: 10.1128/AAC.02682-15
Contribution of Gag and Protease to HIV-1 Phenotypic Drug Resistance in Pediatric Patients Failing Protease Inhibitor-Based Therapy
Abstract
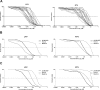
Protease inhibitors (PIs) are used as a first-line regimen in HIV-1-infected children. Here we investigated the phenotypic consequences of amino acid changes in Gag and protease on lopinavir (LPV) and ritonavir (RTV) susceptibility among pediatric patients failing PI therapy. The Gag-protease from isolates from 20 HIV-1 subtype C-infected pediatric patients failing an LPV and/or RTV-based regimen was phenotyped using a nonreplicativein vitroassay. Changes in sensitivity to LPV and RTV relative to that of the matched baseline (pretherapy) sample were calculated. Gag and protease amino acid substitutions associated with PI failure were created in a reference clone by site-directed mutagenesis and assessed. Predicted phenotypes were determined using the Stanford drug resistance algorithm. Phenotypic resistance or reduced susceptibility to RTV and/or LPV was observed in isolates from 10 (50%) patients, all of whom had been treated with RTV. In most cases, this was associated with protease resistance mutations, but substitutions at Gag cleavage and noncleavage sites were also detected. Gag amino acid substitutions were also found in isolates from three patients with reduced drug susceptibilities who had wild-type protease. Site-directed mutagenesis confirmed that some amino acid changes in Gag contributed to PI resistance but only in the presence of major protease resistance-associated substitutions. The isolates from all patients who received LPV exclusively were phenotypically susceptible. Baseline isolates from the 20 patients showed a large (47-fold) range in the 50% effective concentration of LPV, which accounted for most of the discordance seen between the experimentally determined and the predicted phenotypes. Overall, the inclusion of thegaggene and the use of matched baseline samples provided a more comprehensive assessment of the effect of PI-induced amino acid changes on PI resistance. The lack of phenotypic resistance to LPV supports the continued use of this drug in pediatric patients.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures

Similar articles
-
Evidence for Reduced Drug Susceptibility without Emergence of Major Protease Mutations following Protease Inhibitor Monotherapy Failure in the SARA Trial.PLoS One. 2015 Sep 18;10(9):e0137834. doi: 10.1371/journal.pone.0137834. eCollection 2015. PLoS One. 2015. PMID: 26382239 Free PMC article. Clinical Trial.
-
A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism.PLoS Med. 2007 Jan;4(1):e36. doi: 10.1371/journal.pmed.0040036. PLoS Med. 2007. PMID: 17227139 Free PMC article.
-
Single genome sequencing of HIV-1 gag and protease resistance mutations at virologic failure during the OK04 trial of simplified versus standard maintenance therapy.Antivir Ther. 2011;16(5):725-32. doi: 10.3851/IMP1812. Antivir Ther. 2011. PMID: 21817194 Free PMC article.
-
The influence of protease inhibitor resistance profiles on selection of HIV therapy in treatment-naive patients.Antivir Ther. 2004 Jun;9(3):301-14. Antivir Ther. 2004. PMID: 15259893 Review.
-
Acquisition of multi-PI (protease inhibitor) resistance in HIV-1 in vivo and in vitro.Curr Pharm Des. 2004;10(32):4055-64. doi: 10.2174/1381612043382477. Curr Pharm Des. 2004. PMID: 15579087 Review.
Cited by
-
Gag-protease coevolution analyses define novel structural surfaces in the HIV-1 matrix and capsid involved in resistance to Protease Inhibitors.Sci Rep. 2017 Jun 16;7(1):3717. doi: 10.1038/s41598-017-03260-4. Sci Rep. 2017. PMID: 28623276 Free PMC article.
-
Gp41 and Gag amino acids linked to HIV-1 protease inhibitor-based second-line failure in HIV-1 subtype A from Western Kenya.J Int AIDS Soc. 2017 Nov;20(3):e25024. doi: 10.1002/jia2.25024. J Int AIDS Soc. 2017. PMID: 29098809 Free PMC article.
-
Baseline PI susceptibility by HIV-1 Gag-protease phenotyping and subsequent virological suppression with PI-based second-line ART in Nigeria.J Antimicrob Chemother. 2019 May 1;74(5):1402-1407. doi: 10.1093/jac/dkz005. J Antimicrob Chemother. 2019. PMID: 30726945 Free PMC article.
-
Wide variation in susceptibility of transmitted/founder HIV-1 subtype C Isolates to protease inhibitors and association with in vitro replication efficiency.Sci Rep. 2016 Nov 30;6:38153. doi: 10.1038/srep38153. Sci Rep. 2016. PMID: 27901085 Free PMC article.
-
In Vivo Emergence of a Novel Protease Inhibitor Resistance Signature in HIV-1 Matrix.mBio. 2020 Nov 3;11(6):e02036-20. doi: 10.1128/mBio.02036-20. mBio. 2020. PMID: 33144375 Free PMC article.
References
-
- Department of Health. 2013. The South African antiretroviral treatment guidelines. Department of Health, Pretoria, South Africa.
-
- WHO. 2013. Global update on HIV treatment 2013: results, impact and opportunities. WHO, Geneva, Switzerland.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

