Whole genome sequencing revealed host adaptation-focused genomic plasticity of pathogenic Leptospira
- PMID: 26833181
- PMCID: PMC4735792
- DOI: 10.1038/srep20020
Whole genome sequencing revealed host adaptation-focused genomic plasticity of pathogenic Leptospira
Abstract
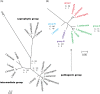
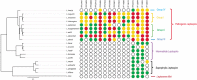
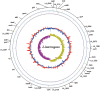
Leptospirosis, caused by pathogenic Leptospira spp., has recently been recognized as an emerging infectious disease worldwide. Despite its severity and global importance, knowledge about the molecular pathogenesis and virulence evolution of Leptospira spp. remains limited. Here we sequenced and analyzed 102 isolates representing global sources. A high genomic variability were observed among different Leptospira species, which was attributed to massive gene gain and loss events allowing for adaptation to specific niche conditions and changing host environments. Horizontal gene transfer and gene duplication allowed the stepwise acquisition of virulence factors in pathogenic Leptospira evolved from a recent common ancestor. More importantly, the abundant expansion of specific virulence-related protein families, such as metalloproteases-associated paralogs, were exclusively identified in pathogenic species, reflecting the importance of these protein families in the pathogenesis of leptospirosis. Our observations also indicated that positive selection played a crucial role on this bacteria adaptation to hosts. These novel findings may lead to greater understanding of the global diversity and virulence evolution of Leptospira spp.
Figures


References
-
- Bharti A. R. et al.. Leptospirosis: a zoonotic disease of global importance. Lancet Infect Dis 3, 757–771 (2003). - PubMed
-
- World Health Organization. Report of the Second Meeting of the Leptospirosis Burden Epidemiology Reference Group. (2010) Available at: http://apps.who.int/iris/bitstream/10665/44588/1/9789241501521_eng.pdf (Accessed: 6th August 2015).
-
- Brenner D. J. et al.. Further determination of DNA relatedness between serogroups and serovars in the family Leptospiraceae with a proposal for Leptospira alexanderi sp. nov. and four new Leptospira genomospecies. Int J Syst Bacteriol 49 Pt 2, 839–858 (1999). - PubMed
-
- Petersen A. M., Boye K., Blom J., Schlichting P. & Krogfelt K. A. First isolation of Leptospira fainei serovar Hurstbridge from two human patients with Weil’s syndrome. J Med Microbiol 50, 96–100 (2001). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

