Comparative fiber property and transcriptome analyses reveal key genes potentially related to high fiber strength in cotton (Gossypium hirsutum L.) line MD52ne
- PMID: 26833213
- PMCID: PMC4736178
- DOI: 10.1186/s12870-016-0727-2
Comparative fiber property and transcriptome analyses reveal key genes potentially related to high fiber strength in cotton (Gossypium hirsutum L.) line MD52ne
Abstract
Background: Individual fiber strength is an important quality attribute that greatly influences the strength of the yarn spun from cotton fibers. Fiber strength is usually measured from bundles of fibers due to the difficulty of reliably measuring strength from individual cotton fibers. However, bundle fiber strength (BFS) is not always correlated with yarn strength since it is affected by multiple fiber properties involved in fiber-to-fiber interactions within a bundle in addition to the individual fiber strength. Molecular mechanisms responsible for regulating individual fiber strength remain unknown. Gossypium hirsutum near isogenic lines (NILs), MD52ne and MD90ne showing variations in BFS provide an opportunity for dissecting the regulatory mechanisms involved in individual fiber strength.
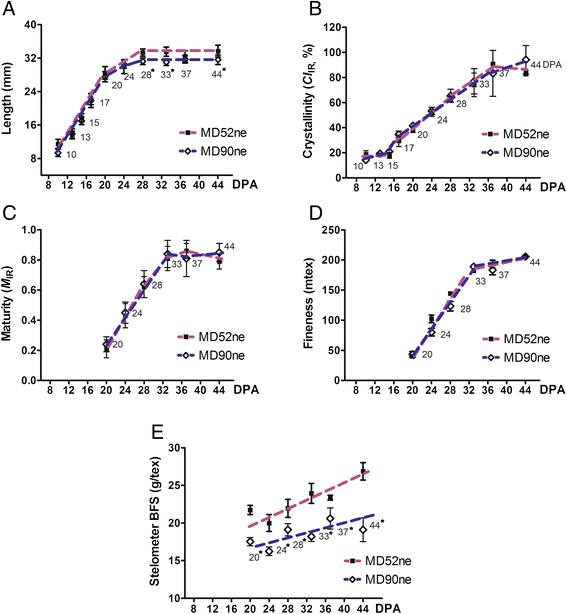
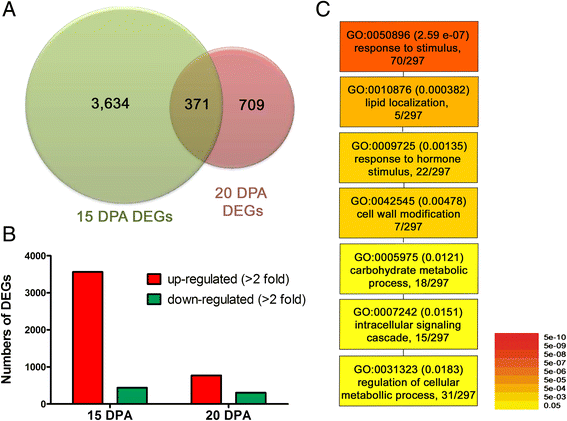
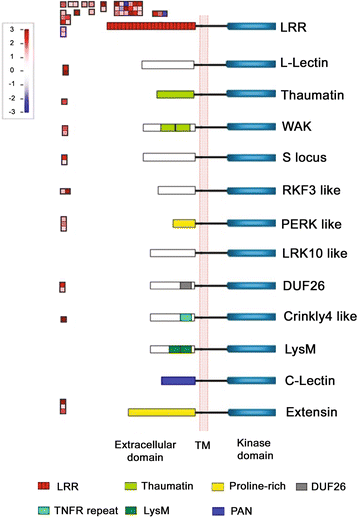
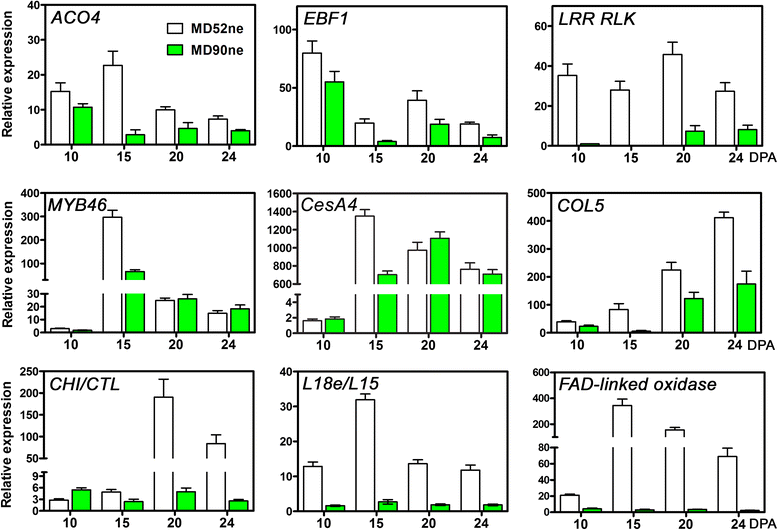
Results: Comprehensive fiber property analyses of the NILs revealed that the superior bundle strength of MD52ne fibers resulted from high individual fiber strength with minor contributions from greater fiber length. Comparative transcriptome analyses of the NILs showed that the superior bundle strength of MD52ne fibers was potentially related to two signaling pathways: one is ethylene and the interconnected phytohormonal pathways that are involved in cotton fiber elongation, and the other is receptor-like kinases (RLKs) signaling pathways that are involved in maintaining cell wall integrity. Multiple RLKs were differentially expressed in MD52ne fibers and localized in genomic regions encompassing the strength quantitative trait loci (QTLs). Several candidate genes involved in crystalline cellulose assembly were also up-regulated in MD52ne fibers while the secondary cell wall was produced.
Conclusion: Comparative phenotypic and transcriptomic analyses revealed differential expressions of the genes involved in crystalline cellulose assembly, ethylene and RLK signaling pathways between the MD52ne and MD90ne developing fibers. Ethylene and its phytohormonal network might promote the elongation of MD52ne fibers and indirectly contribute to the bundle strength by potentially improving fiber-to-fiber interactions. RLKs that were suggested to mediate a coordination of cell elongation and SCW biosynthesis in other plants might be candidate genes for regulating cotton fiber cell wall assembly and strength.
Figures




Similar articles
-
Mapping by sequencing in cotton (Gossypium hirsutum) line MD52ne identified candidate genes for fiber strength and its related quality attributes.Theor Appl Genet. 2016 Jun;129(6):1071-86. doi: 10.1007/s00122-016-2684-4. Epub 2016 Feb 16. Theor Appl Genet. 2016. PMID: 26883043
-
Near-isogenic cotton germplasm lines that differ in fiber-bundle strength have temporal differences in fiber gene expression patterns as revealed by comparative high-throughput profiling.Theor Appl Genet. 2010 May;120(7):1347-66. doi: 10.1007/s00122-010-1260-6. Epub 2010 Jan 20. Theor Appl Genet. 2010. PMID: 20087569
-
Cloning and characterization of homeologous cellulose synthase catalytic subunit 2 genes from allotetraploid cotton (Gossypium hirsutum L.).Gene. 2012 Feb 25;494(2):181-9. doi: 10.1016/j.gene.2011.12.018. Epub 2011 Dec 20. Gene. 2012. PMID: 22200568
-
Dynamic roles and intricate mechanisms of ethylene in epidermal hair development in Arabidopsis and cotton.New Phytol. 2022 Apr;234(2):375-391. doi: 10.1111/nph.17901. Epub 2021 Dec 30. New Phytol. 2022. PMID: 34882809 Review.
-
A comprehensive overview of cotton genomics, biotechnology and molecular biological studies.Sci China Life Sci. 2023 Oct;66(10):2214-2256. doi: 10.1007/s11427-022-2278-0. Epub 2023 Mar 6. Sci China Life Sci. 2023. PMID: 36899210 Review.
Cited by
-
Examining two sets of introgression lines across multiple environments reveals background-independent and stably expressed quantitative trait loci of fiber quality in cotton.Theor Appl Genet. 2020 Jul;133(7):2075-2093. doi: 10.1007/s00122-020-03578-0. Epub 2020 Mar 17. Theor Appl Genet. 2020. PMID: 32185421 Free PMC article.
-
Comparative Transcriptomic Analysis to Identify the Genes Related to Delayed Gland Morphogenesis in Gossypium bickii.Genes (Basel). 2020 Apr 26;11(5):472. doi: 10.3390/genes11050472. Genes (Basel). 2020. PMID: 32357512 Free PMC article.
-
A high-resolution model of gene expression during Gossypium hirsutum (cotton) fiber development.BMC Genomics. 2025 Mar 6;26(1):221. doi: 10.1186/s12864-025-11360-z. BMC Genomics. 2025. PMID: 40050725 Free PMC article.
-
Identification of candidate genes for fiber length quantitative trait loci through RNA-Seq and linkage and physical mapping in cotton.BMC Genomics. 2017 May 31;18(1):427. doi: 10.1186/s12864-017-3812-5. BMC Genomics. 2017. PMID: 28569138 Free PMC article.
-
Mapping by sequencing in cotton (Gossypium hirsutum) line MD52ne identified candidate genes for fiber strength and its related quality attributes.Theor Appl Genet. 2016 Jun;129(6):1071-86. doi: 10.1007/s00122-016-2684-4. Epub 2016 Feb 16. Theor Appl Genet. 2016. PMID: 26883043
References
-
- Wakelyn PJ, Bertoniere NR, French AD, Thibodeaux DP, Triplett BA, Rousselle M-A, et al. Cotton fiber chemistry and technology, vol. 17. New York, USA: CRC Press; 2010.
-
- Nichols N, Martin V, Devine J, Li H, Jones D, Hake K. Variety performance: a critical issue for cotton competitiveness. Raliegh, North Carolina: Cotton Incorporated; 2012.
-
- Haigler C. Physiological and anatomical factors determining fiber structure and utility. In: Physiology of cotton. New York, USA: Springer; 2010. p. 33-47.
-
- Bradow JM, Davidonis GH. Quantitation of fiber quality and the cotton production-processing interface: A physiologist’s perspective. J Cotton Sci. 2000;4:34–64.
-
- Frydrych I, Thibodeaux DP. Fiber quality evaluation-current and future trends/ intrinsic value of fiber quality in cotton. In: Wakelyn PJ, Chaudhry MR, editors. Cotton: technology for the 21st century. Washington DC: International Cotton Advisory Committee; 2010. pp. 251–296.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

