Molecular characterization of human coronaviruses and their circulation dynamics in Kenya, 2009-2012
- PMID: 26833249
- PMCID: PMC4736488
- DOI: 10.1186/s12985-016-0474-x
Molecular characterization of human coronaviruses and their circulation dynamics in Kenya, 2009-2012
Abstract
Background: Human Coronaviruses (HCoV) are a common cause of respiratory illnesses and are responsible for considerable morbidity and hospitalization across all age groups especially in individuals with compromised immunity. There are six known species of HCoV: HCoV-229E, HCoV-NL63, HCoV-HKU1, HCoV-OC43, MERS-CoV and SARS-HCoV. Although studies have shown evidence of global distribution of HCoVs, there is limited information on their presence and distribution in Kenya.
Methods: HCoV strains that circulated in Kenya were retrospectively diagnosed and molecularly characterized. A total of 417 nasopharyngeal specimens obtained between January 2009 and December 2012 from around Kenya were analyzed by a real time RT-PCR using HCoV-specific primers. HCoV-positive specimens were subsequently inoculated onto monolayers of LL-CMK2 cells. The isolated viruses were characterized by RT-PCR amplification and sequencing of the partial polymerase (pol) gene.
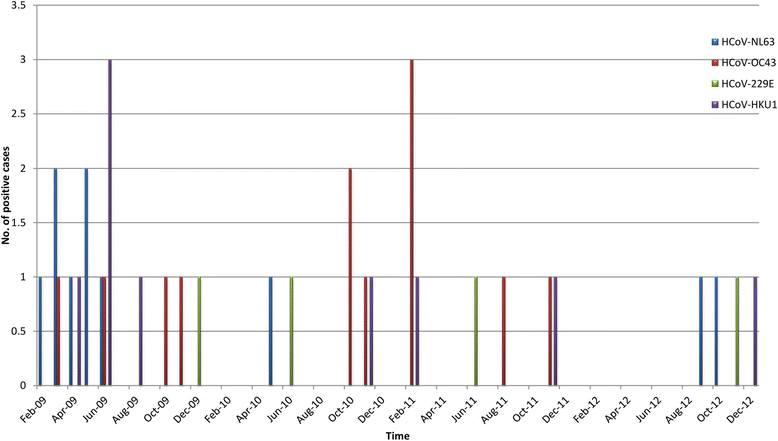
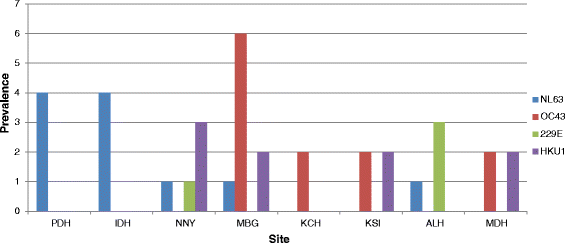
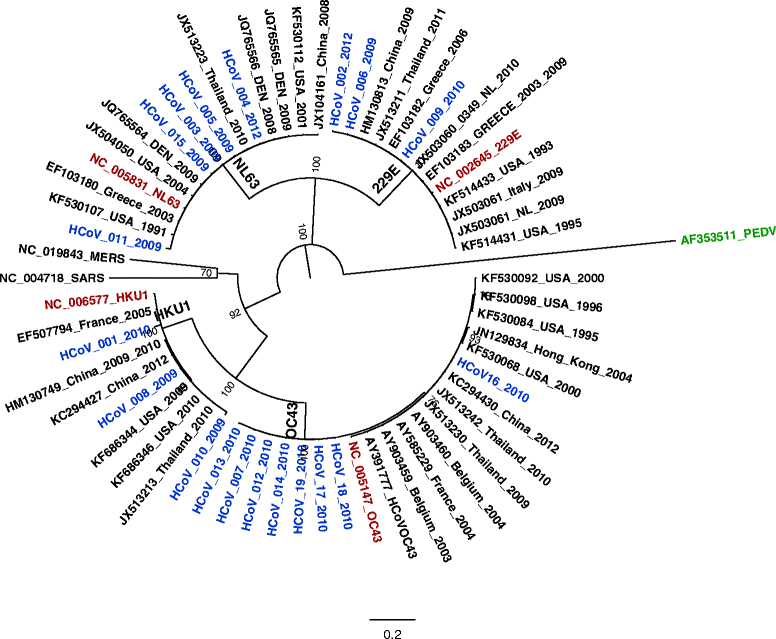
Results: The prevalence of HCoV infection was as follows: out of the 417 specimens, 35 (8.4 %) were positive for HCoV, comprising 10 (2.4 %) HCoV-NL63, 12 (2.9 %) HCoV-OC43, 9 (2.1 %) HCoV-HKU1, and 4 (1 %) HCoV-229E. The Kenyan HCoV strains displayed high sequence homology to the prototypes and contemporaneous strains. Evolution analysis showed that the Kenyan HCoV-OC43 and HCoV-NL63 isolates were under purifying selection. Phylogenetic evolutionary analyses confirmed the identities of three HCoV-HKU1, five HCoV-NL63, eight HCoV-OC43 and three HCoV-229E.
Conclusions: There were yearly variations in the prevalence and circulation patterns of individual HCoVs in Kenya. This paper reports on the first molecular characterization of human Coronaviruses in Kenya, which play an important role in causing acute respiratory infections among children.
Figures
References
-
- Xu HF, Wang M, Zhang ZB, Zou XZ, Gao Y, Liu XN, et al. An epidemiologic investigation on infection with severe acute respiratory syndrome coronavirus in wild animals traders in Guangzhou. Zhonghua Yu Fang Yi Xue Za Zhi. 2004;38(2):81–3. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

