Mito-priming as a method to engineer Bcl-2 addiction
- PMID: 26833356
- PMCID: PMC4740867
- DOI: 10.1038/ncomms10538
Mito-priming as a method to engineer Bcl-2 addiction
Abstract
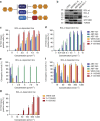
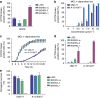
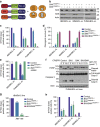
Most apoptotic stimuli require mitochondrial outer membrane permeabilization (MOMP) in order to execute cell death. As such, MOMP is subject to tight control by Bcl-2 family proteins. We have developed a powerful new technique to investigate Bcl-2-mediated regulation of MOMP. This method, called mito-priming, uses co-expression of pro- and anti-apoptotic Bcl-2 proteins to engineer Bcl-2 addiction. On addition of Bcl-2 targeting BH3 mimetics, mito-primed cells undergo apoptosis in a rapid and synchronous manner. Using this method we have comprehensively surveyed the efficacy of BH3 mimetic compounds, identifying potent and specific MCL-1 inhibitors. Furthermore, by combining different pro- and anti-apoptotic Bcl-2 pairings together with CRISPR/Cas9-based genome editing, we find that tBID and PUMA can preferentially kill in a BAK-dependent manner. In summary, mito-priming represents a facile and robust means to trigger mitochondrial apoptosis.
Figures



References
-
- Tait S. W. & Green D. R. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 11, 621–632 (2010). - PubMed
-
- Czabotar P. E., Lessene G., Strasser A. & Adams J. M. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 15, 49–63 (2014). - PubMed
-
- Hanahan D. & Weinberg R. A. The hallmarks of cancer. Cell 100, 57–70 (2000). - PubMed
-
- Certo M. et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer cell 9, 351–365 (2006). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

