MLL1 is essential for the senescence-associated secretory phenotype
- PMID: 26833731
- PMCID: PMC4743061
- DOI: 10.1101/gad.271882.115
MLL1 is essential for the senescence-associated secretory phenotype
Abstract
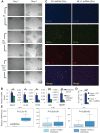
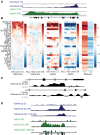
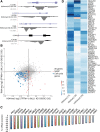
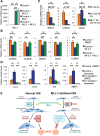
Oncogene-induced senescence (OIS) and therapy-induced senescence (TIS), while tumor-suppressive, also promote procarcinogenic effects by activating the DNA damage response (DDR), which in turn induces inflammation. This inflammatory response prominently includes an array of cytokines known as the senescence-associated secretory phenotype (SASP). Previous observations link the transcription-associated methyltransferase and oncoprotein MLL1 to the DDR, leading us to investigate the role of MLL1 in SASP expression. Our findings reveal direct MLL1 epigenetic control over proproliferative cell cycle genes: MLL1 inhibition represses expression of proproliferative cell cycle regulators required for DNA replication and DDR activation, thus disabling SASP expression. Strikingly, however, these effects of MLL1 inhibition on SASP gene expression do not impair OIS and, furthermore, abolish the ability of the SASP to enhance cancer cell proliferation. More broadly, MLL1 inhibition also reduces "SASP-like" inflammatory gene expression from cancer cells in vitro and in vivo independently of senescence. Taken together, these data demonstrate that MLL1 inhibition may be a powerful and effective strategy for inducing cancerous growth arrest through the direct epigenetic regulation of proliferation-promoting genes and the avoidance of deleterious OIS- or TIS-related tumor secretomes, which can promote both drug resistance and tumor progression.
Keywords: DNA damage response; MLL1; epigenetic; inflammation; oncogene-induced senescence; senescence-associated secretory phenotype.
© 2016 Capell et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



Similar articles
-
Tumorigenicity of Ewing sarcoma is critically dependent on the trithorax proteins MLL1 and menin.Oncotarget. 2017 Jan 3;8(1):458-471. doi: 10.18632/oncotarget.13444. Oncotarget. 2017. PMID: 27888797 Free PMC article.
-
Targeting MLL1/WDR5-Mediated Epigenetic Regulation Mitigates Peritoneal Fibrosis by Reducing p16INK4a.FASEB J. 2025 Apr 30;39(8):e70543. doi: 10.1096/fj.202402382R. FASEB J. 2025. PMID: 40232893
-
The STAT4/MLL1 Epigenetic Axis Regulates the Antimicrobial Functions of Murine Macrophages.J Immunol. 2017 Sep 1;199(5):1865-1874. doi: 10.4049/jimmunol.1601272. Epub 2017 Jul 21. J Immunol. 2017. PMID: 28733487 Free PMC article.
-
Keeping the senescence secretome under control: Molecular reins on the senescence-associated secretory phenotype.Exp Gerontol. 2016 Sep;82:39-49. doi: 10.1016/j.exger.2016.05.010. Epub 2016 May 25. Exp Gerontol. 2016. PMID: 27235851 Review.
-
Emerging role of NF-κB signaling in the induction of senescence-associated secretory phenotype (SASP).Cell Signal. 2012 Apr;24(4):835-45. doi: 10.1016/j.cellsig.2011.12.006. Epub 2011 Dec 11. Cell Signal. 2012. PMID: 22182507 Review.
Cited by
-
The role of the dynamic epigenetic landscape in senescence: orchestrating SASP expression.NPJ Aging. 2024 Oct 24;10(1):48. doi: 10.1038/s41514-024-00172-2. NPJ Aging. 2024. PMID: 39448585 Free PMC article. Review.
-
Intracellular signaling modules linking DNA damage to secretome changes in senescent melanoma cells.Melanoma Res. 2020 Aug;30(4):336-347. doi: 10.1097/CMR.0000000000000671. Melanoma Res. 2020. PMID: 32628430 Free PMC article.
-
Astrocyte Senescence and Alzheimer's Disease: A Review.Front Aging Neurosci. 2020 Jun 9;12:148. doi: 10.3389/fnagi.2020.00148. eCollection 2020. Front Aging Neurosci. 2020. PMID: 32581763 Free PMC article. Review.
-
Uncoupling the Senescence-Associated Secretory Phenotype from Cell Cycle Exit via Interleukin-1 Inactivation Unveils Its Protumorigenic Role.Mol Cell Biol. 2019 May 28;39(12):e00586-18. doi: 10.1128/MCB.00586-18. Print 2019 Jun 15. Mol Cell Biol. 2019. PMID: 30988157 Free PMC article.
-
Flavonoids: Broad Spectrum Agents on Chronic Inflammation.Biomol Ther (Seoul). 2019 May 1;27(3):241-253. doi: 10.4062/biomolther.2019.034. Biomol Ther (Seoul). 2019. PMID: 31006180 Free PMC article. Review.
References
-
- Algire C, Moiseeva O, Deschenes-Simard X, Amrein L, Petruccelli L, Birman E, Viollet B, Ferbeyre G, Pollak MN. 2012. Metformin reduces endogenous reactive oxygen species and associated DNA damage. Cancer Prev Res 5: 536–543. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
