On the satisfaction of backbone-carbonyl lone pairs of electrons in protein structures
- PMID: 26833776
- PMCID: PMC4941217
- DOI: 10.1002/pro.2896
On the satisfaction of backbone-carbonyl lone pairs of electrons in protein structures
Abstract
Protein structures are stabilized by a variety of noncovalent interactions (NCIs), including the hydrophobic effect, hydrogen bonds, electrostatic forces and van der Waals' interactions. Our knowledge of the contributions of NCIs, and the interplay between them remains incomplete. This has implications for computational modeling of NCIs, and our ability to understand and predict protein structure, stability, and function. One consideration is the satisfaction of the full potential for NCIs made by backbone atoms. Most commonly, backbone-carbonyl oxygen atoms located within α-helices and β-sheets are depicted as making a single hydrogen bond. However, there are two lone pairs of electrons to be satisfied for each of these atoms. To explore this, we used operational geometric definitions to generate an inventory of NCIs for backbone-carbonyl oxygen atoms from a set of high-resolution protein structures and associated molecular-dynamics simulations in water. We included more-recently appreciated, but weaker NCIs in our analysis, such as n→π* interactions, Cα-H bonds and methyl-H bonds. The data demonstrate balanced, dynamic systems for all proteins, with most backbone-carbonyl oxygen atoms being satisfied by two NCIs most of the time. Combinations of NCIs made may correlate with secondary structure type, though in subtly different ways from traditional models of α- and β-structure. In addition, we find examples of under- and over-satisfied carbonyl-oxygen atoms, and we identify both sequence-dependent and sequence-independent secondary-structural motifs in which these reside. Our analysis provides a more-detailed understanding of these contributors to protein structure and stability, which will be of use in protein modeling, engineering and design.
Keywords: bioinformatics; hydrogen bonding; noncovalent interactions; n→π* interactions; protein folding; protein stability; protein structure.
© 2016 The Authors Protein Science published by Wiley Periodicals, Inc. on behalf of The Protein Society.
Figures
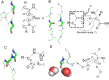
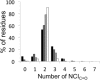
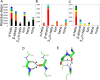
 ), (PDB 1G66, residues A26‐A30). Secondary structures were assigned by Promotif,42 which uses a modified version of the Kabsch and Sander DSSP algorithm.58 Categories “E” and “B” were combined into a single β‐structure category.
), (PDB 1G66, residues A26‐A30). Secondary structures were assigned by Promotif,42 which uses a modified version of the Kabsch and Sander DSSP algorithm.58 Categories “E” and “B” were combined into a single β‐structure category.
 ) and one 1 × CHX (▪). The residue providing the CHx has been truncated for clarity. (B) Motifs at the α‐helical Ntermini with 2 × NHbb (•) plus 1 × n→π* interaction (
) and one 1 × CHX (▪). The residue providing the CHx has been truncated for clarity. (B) Motifs at the α‐helical Ntermini with 2 × NHbb (•) plus 1 × n→π* interaction ( ). (C) α‐helical C‐termini with 1 × OHsc, (▲), 1 × NHbb (•) and 1 × n→π* interaction (
). (C) α‐helical C‐termini with 1 × OHsc, (▲), 1 × NHbb (•) and 1 × n→π* interaction ( ), and associated WebLogos59 indicating the amino‐acid frequencies from sequences in our dataset that display this motif. Structural images prepared with PyMOL (
), and associated WebLogos59 indicating the amino‐acid frequencies from sequences in our dataset that display this motif. Structural images prepared with PyMOL (Similar articles
-
Interplay of hydrogen bonds and n→π* interactions in proteins.J Am Chem Soc. 2013 Dec 11;135(49):18682-8. doi: 10.1021/ja4106122. Epub 2013 Dec 3. J Am Chem Soc. 2013. PMID: 24256417 Free PMC article.
-
Noncovalent interactions in proteins and nucleic acids: beyond hydrogen bonding and π-stacking.Chem Soc Rev. 2022 Jun 6;51(11):4261-4286. doi: 10.1039/d2cs00133k. Chem Soc Rev. 2022. PMID: 35560317 Review.
-
Statistical and molecular dynamics studies of buried waters in globular proteins.Proteins. 2005 Aug 15;60(3):450-63. doi: 10.1002/prot.20511. Proteins. 2005. PMID: 15937899
-
n-->pi* interactions in proteins.Nat Chem Biol. 2010 Aug;6(8):615-20. doi: 10.1038/nchembio.406. Epub 2010 Jul 11. Nat Chem Biol. 2010. PMID: 20622857 Free PMC article.
-
Roles of electrostatic interaction in proteins.Q Rev Biophys. 1996 Feb;29(1):1-90. doi: 10.1017/s0033583500005746. Q Rev Biophys. 1996. PMID: 8783394 Review.
Cited by
-
Secondary Forces in Protein Folding.ACS Chem Biol. 2019 Aug 16;14(8):1677-1686. doi: 10.1021/acschembio.9b00339. Epub 2019 Jun 19. ACS Chem Biol. 2019. PMID: 31243961 Free PMC article. Review.
-
The n→π* Interaction.Acc Chem Res. 2017 Aug 15;50(8):1838-1846. doi: 10.1021/acs.accounts.7b00121. Epub 2017 Jul 23. Acc Chem Res. 2017. PMID: 28735540 Free PMC article.
-
When are two hydrogen bonds better than one? Accurate first-principles models explain the balance of hydrogen bond donors and acceptors found in proteins.Chem Sci. 2020 Nov 19;12(3):1147-1162. doi: 10.1039/d0sc05084a. eCollection 2021 Jan 21. Chem Sci. 2020. PMID: 35382134 Free PMC article.
-
Side chain to main chain hydrogen bonds stabilize a polyglutamine helix in a transcription factor.Nat Commun. 2019 May 2;10(1):2034. doi: 10.1038/s41467-019-09923-2. Nat Commun. 2019. PMID: 31048691 Free PMC article.
-
A prevalent intraresidue hydrogen bond stabilizes proteins.Nat Chem Biol. 2016 Dec;12(12):1084-1088. doi: 10.1038/nchembio.2206. Epub 2016 Oct 17. Nat Chem Biol. 2016. PMID: 27748749 Free PMC article.
References
-
- Anfinsen CB (1973) Principles that govern the folding of protein chains. Science 181:223–230. - PubMed
-
- Dill K (1990) Dominant forces in protein folding. Biochemistry 29:7133–7155. - PubMed
-
- Derewenda ZS, Lee L, Derewenda U (1995) The occurrence of C‐H…O hydrogen bonds in proteins. J Mol Biol 252:248–262. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

