Unraveling the Enzymatic Basis of Wine "Flavorome": A Phylo-Functional Study of Wine Related Yeast Species
- PMID: 26834730
- PMCID: PMC4718978
- DOI: 10.3389/fmicb.2016.00012
Unraveling the Enzymatic Basis of Wine "Flavorome": A Phylo-Functional Study of Wine Related Yeast Species
Abstract
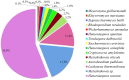
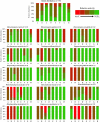
Non-Saccharomyces yeasts are a heterogeneous microbial group involved in the early stages of wine fermentation. The high enzymatic potential of these yeasts makes them a useful tool for increasing the final organoleptic characteristics of wines in spite of their low fermentative power. Their physiology and contribution to wine quality are still poorly understood, with most current knowledge being acquired empirically and in most cases based in single species and strains. This work analyzed the metabolic potential of 770 yeast isolates from different enological origins and representing 15 different species, by studying their production of enzymes of enological interest and linking phylogenetic and enzymatic data. The isolates were screened for glycosidase enzymes related to terpene aroma release, the β-lyase activity responsible for the release of volatile thiols, and sulfite reductase. Apart from these aroma-related activities, protease, polygalacturonase and cellulase activities were also studied in the entire yeast collection, being related to the improvement of different technological and sensorial features of wines. In this context, and in terms of abundance, two different groups were established, with α-L-arabinofuranosidase, polygalacturonase and cellulase being the less abundant activities. By contrast, β-glucosidase and protease activities were widespread in the yeast collection studied. A classical phylogenetic study involving the partial sequencing of 26S rDNA was conducted in conjunction with the enzymatic profiles of the 770 yeast isolates for further typing, complementing the phylogenetic relationships established by using 26S rDNA. This has rendered it possible to foresee the contribution different yeast species make to wine quality and their potential applicability as pure inocula, establishing species-specific behavior. These consistent results allowed us to design future targeted studies on the impact different non-Saccharomyces yeast species have on wine quality, understanding intra and interspecific enzymatic odds and, therefore, aiming to predict the most suitable application for the current non-Saccharomyces strains, as well as the potential future applications of new strains. This work therefore contributes to a better understanding of the concept of wine microbiome and its potential consequences for wine quality, as well as to the knowledge of non-Saccharomyces yeasts for their use in the wine industry.
Keywords: enological enzymes; microbial terroir; non-Saccharomyces; phylo-functional study; targeted yeast selection.
Figures


Similar articles
-
Occurrence and enological properties of two new non-conventional yeasts (Nakazawaea ishiwadae and Lodderomyces elongisporus) in wine fermentations.Int J Food Microbiol. 2019 Sep 16;305:108255. doi: 10.1016/j.ijfoodmicro.2019.108255. Epub 2019 Jun 20. Int J Food Microbiol. 2019. PMID: 31252247
-
Non-Saccharomyces Yeasts from Organic Vineyards as Spontaneous Fermentation Agents.Foods. 2023 Oct 2;12(19):3644. doi: 10.3390/foods12193644. Foods. 2023. PMID: 37835297 Free PMC article.
-
Partial 26S rDNA restriction analysis as a tool to characterise non-Saccharomyces yeasts present during red wine fermentations.Int J Food Microbiol. 2005 Jun 25;102(1):49-56. doi: 10.1016/j.ijfoodmicro.2005.01.005. Int J Food Microbiol. 2005. PMID: 15925001
-
The Important Contribution of Non-Saccharomyces Yeasts to the Aroma Complexity of Wine: A Review.Foods. 2020 Dec 23;10(1):13. doi: 10.3390/foods10010013. Foods. 2020. PMID: 33374550 Free PMC article. Review.
-
Past and Future of Non-Saccharomyces Yeasts: From Spoilage Microorganisms to Biotechnological Tools for Improving Wine Aroma Complexity.Front Microbiol. 2016 Mar 31;7:411. doi: 10.3389/fmicb.2016.00411. eCollection 2016. Front Microbiol. 2016. PMID: 27065975 Free PMC article. Review.
Cited by
-
Metataxonomic Analysis of Grape Microbiota During Wine Fermentation Reveals the Distinction of Cyprus Regional terroirs.Front Microbiol. 2021 Sep 22;12:726483. doi: 10.3389/fmicb.2021.726483. eCollection 2021. Front Microbiol. 2021. PMID: 34630353 Free PMC article.
-
Diversity of Mycobiota in Spanish Grape Berries and Selection of Hanseniaspora uvarum U1 to Prevent Mycotoxin Contamination.Toxins (Basel). 2021 Sep 13;13(9):649. doi: 10.3390/toxins13090649. Toxins (Basel). 2021. PMID: 34564653 Free PMC article.
-
Techniques for Dealcoholization of Wines: Their Impact on Wine Phenolic Composition, Volatile Composition, and Sensory Characteristics.Foods. 2021 Oct 18;10(10):2498. doi: 10.3390/foods10102498. Foods. 2021. PMID: 34681547 Free PMC article. Review.
-
Genome Sequencing and Comparative Analysis of Three Hanseniaspora uvarum Indigenous Wine Strains Reveal Remarkable Biotechnological Potential.Front Microbiol. 2020 Jan 21;10:3133. doi: 10.3389/fmicb.2019.03133. eCollection 2019. Front Microbiol. 2020. PMID: 32038567 Free PMC article.
-
Strong ethanol- and frequency-dependent ecological interactions in a community of wine-fermenting yeasts.Commun Biol. 2023 Sep 13;6(1):939. doi: 10.1038/s42003-023-05284-1. Commun Biol. 2023. PMID: 37704781 Free PMC article.
References
-
- Benito A., Calderón F., Palomero F., Benito S. (2015). Combined use of selected Schizosaccharomyces pombe and Lachancea thermotolerans yeast strains as an alternative to the traditional malolactic fermentation in red wine production. Molecules 20, 9510–9523. 10.3390/molecules20069510 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

