Tumor Suppressor DLEC1 can Stimulate the Proliferation of Cancer Cells When AP-2ɑ2 is Down-Regulated in HCT116
- PMID: 26834787
- PMCID: PMC4723729
- DOI: 10.5812/hepatmon.29829
Tumor Suppressor DLEC1 can Stimulate the Proliferation of Cancer Cells When AP-2ɑ2 is Down-Regulated in HCT116
Abstract
Background: The molecular mechanisms of tumor suppressor gene DLEC1 are largely unknown.
Objectives: In this study, we established DLEC1 over-expression stable clones to study the cellular function of DLEC1 in the colorectal cancer cell line, HCT116.
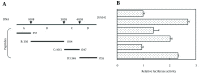
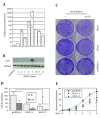
Materials and methods: Stable clones with DLEC1 over-expression were first established by the transfection of DLEC1 expression construct pcDNA31DLEC1 in HCT116. On G418 selection, positive stable clones were screened for DLEC1 expression level by conventional reverse transcription-polymerase chain reaction (RT-PCR), and verified by real-time RT-PCR and Western blotting. Subsequently, these stable clones were subjected to colony formation and cell cycle analyses and identification of factors involved in G1 arrest. Lastly, three stable clones, DLEC1-7 (highest DLEC1 expression), DLEC1-3 (lowest expression) and pcDNA31 vector control, were employed to analyze cell proliferation and cell cycle after AP-2α2 knockdown by siRNAs.
Results: The DLEC1 over-expression was found to reduce the number of colonies in colony formation and to induce G1 arrest in seven clones, and apoptosis in one clone in the cell cycle analysis. Furthermore, regardless of the different cell cycle defects in all eight stable clones, the expression level of transcriptional factor AP-2α2 was found to be elevated. More interestingly, we found that when AP-2α2 was knocked down, DLEC1 over-expression neither suppressed cancer cell growth nor induced G1 arrest, yet, instead promoted cell growth and decreased cells in the G1 fraction. This promotion of cell proliferation and release of G1 cells also seemed to be proportional to DLEC1 expression levels in DLEC1 stable clones.
Conclusions: DLEC1 suppresses tumor cell growth the presence of AP-2α2 and stimulates cell proliferation in the down-regulation of AP-2α2 in DLEC1 over-expression stable clones of HTC116.
Keywords: AP-2ɑ; Cell Proliferation; DLEC1; Human; Promotion.
Figures



Similar articles
-
The pro-survival function of DLEC1 and its protection of cancer cells against 5-FU-induced apoptosis through up-regulation of BCL-XL.Cytotechnology. 2019 Feb;71(1):23-33. doi: 10.1007/s10616-018-0258-9. Epub 2019 Jan 3. Cytotechnology. 2019. PMID: 30607648 Free PMC article.
-
The tumor suppressor gene DLEC1 is frequently silenced by DNA methylation in hepatocellular carcinoma and induces G1 arrest in cell cycle.J Hepatol. 2008 Mar;48(3):433-41. doi: 10.1016/j.jhep.2007.11.015. Epub 2007 Dec 31. J Hepatol. 2008. PMID: 18191269
-
Curcumin inhibits anchorage-independent growth of HT29 human colon cancer cells by targeting epigenetic restoration of the tumor suppressor gene DLEC1.Biochem Pharmacol. 2015 Mar 15;94(2):69-78. doi: 10.1016/j.bcp.2015.01.009. Epub 2015 Jan 29. Biochem Pharmacol. 2015. PMID: 25640947 Free PMC article.
-
DLEC1, a 3p tumor suppressor, represses NF-κB signaling and is methylated in prostate cancer.J Mol Med (Berl). 2015 Jun;93(6):691-701. doi: 10.1007/s00109-015-1255-5. Epub 2015 Feb 5. J Mol Med (Berl). 2015. PMID: 25648635
-
Wogonin induced G1 cell cycle arrest by regulating Wnt/β-catenin signaling pathway and inactivating CDK8 in human colorectal cancer carcinoma cells.Toxicology. 2013 Oct 4;312:36-47. doi: 10.1016/j.tox.2013.07.013. Epub 2013 Jul 30. Toxicology. 2013. PMID: 23907061 Review.
Cited by
-
The pro-survival function of DLEC1 and its protection of cancer cells against 5-FU-induced apoptosis through up-regulation of BCL-XL.Cytotechnology. 2019 Feb;71(1):23-33. doi: 10.1007/s10616-018-0258-9. Epub 2019 Jan 3. Cytotechnology. 2019. PMID: 30607648 Free PMC article.
References
-
- Daigo Y, Nishiwaki T, Kawasoe T, Tamari M, Tsuchiya E, Nakamura Y. Molecular cloning of a candidate tumor suppressor gene, DLC1, from chromosome 3p21.3. Cancer Res. 1999;59(8):1966–72. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
