MetFrag relaunched: incorporating strategies beyond in silico fragmentation
- PMID: 26834843
- PMCID: PMC4732001
- DOI: 10.1186/s13321-016-0115-9
MetFrag relaunched: incorporating strategies beyond in silico fragmentation
Abstract
Background: The in silico fragmenter MetFrag, launched in 2010, was one of the first approaches combining compound database searching and fragmentation prediction for small molecule identification from tandem mass spectrometry data. Since then many new approaches have evolved, as has MetFrag itself. This article details the latest developments to MetFrag and its use in small molecule identification since the original publication.
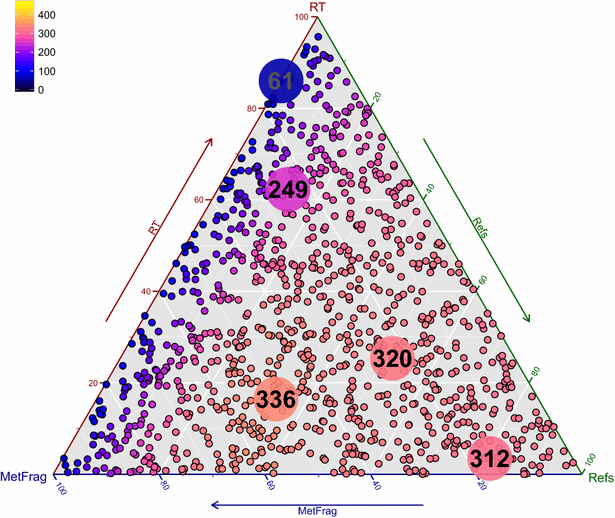
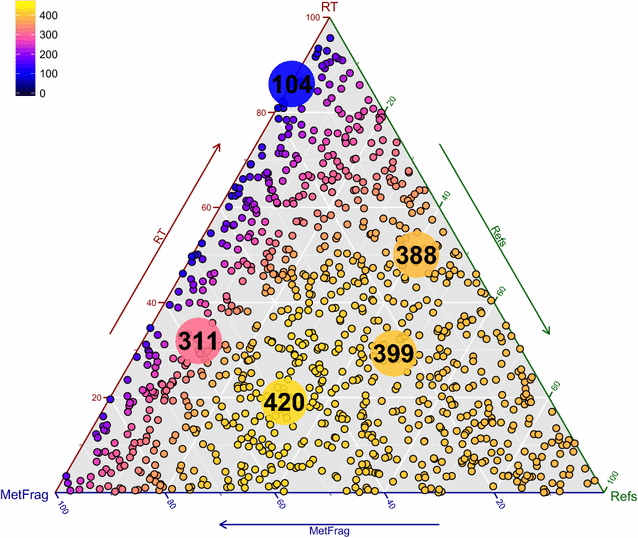
Results: MetFrag has gone through algorithmic and scoring refinements. New features include the retrieval of reference, data source and patent information via ChemSpider and PubChem web services, as well as InChIKey filtering to reduce candidate redundancy due to stereoisomerism. Candidates can be filtered or scored differently based on criteria like occurence of certain elements and/or substructures prior to fragmentation, or presence in so-called "suspect lists". Retention time information can now be calculated either within MetFrag with a sufficient amount of user-provided retention times, or incorporated separately as "user-defined scores" to be included in candidate ranking. The changes to MetFrag were evaluated on the original dataset as well as a dataset of 473 merged high resolution tandem mass spectra (HR-MS/MS) and compared with another open source in silico fragmenter, CFM-ID. Using HR-MS/MS information only, MetFrag2.2 and CFM-ID had 30 and 43 Top 1 ranks, respectively, using PubChem as a database. Including reference and retention information in MetFrag2.2 improved this to 420 and 336 Top 1 ranks with ChemSpider and PubChem (89 and 71 %), respectively, and even up to 343 Top 1 ranks (PubChem) when combining with CFM-ID. The optimal parameters and weights were verified using three additional datasets of 824 merged HR-MS/MS spectra in total. Further examples are given to demonstrate flexibility of the enhanced features.
Conclusions: In many cases additional information is available from the experimental context to add to small molecule identification, which is especially useful where the mass spectrum alone is not sufficient for candidate selection from a large number of candidates. The results achieved with MetFrag2.2 clearly show the benefit of considering this additional information. The new functions greatly enhance the chance of identification success and have been incorporated into a command line interface in a flexible way designed to be integrated into high throughput workflows. Feedback on the command line version of MetFrag2.2 available at http://c-ruttkies.github.io/MetFrag/ is welcome.
Keywords: Compound identification; High resolution mass spectrometry; In silico fragmentation; Metabolomics; Structure elucidation.
Figures
Similar articles
-
Supporting non-target identification by adding hydrogen deuterium exchange MS/MS capabilities to MetFrag.Anal Bioanal Chem. 2019 Jul;411(19):4683-4700. doi: 10.1007/s00216-019-01885-0. Epub 2019 Jun 17. Anal Bioanal Chem. 2019. PMID: 31209548 Free PMC article.
-
In silico fragmentation for computer assisted identification of metabolite mass spectra.BMC Bioinformatics. 2010 Mar 22;11:148. doi: 10.1186/1471-2105-11-148. BMC Bioinformatics. 2010. PMID: 20307295 Free PMC article.
-
Annotation of metabolites from gas chromatography/atmospheric pressure chemical ionization tandem mass spectrometry data using an in silico generated compound database and MetFrag.Rapid Commun Mass Spectrom. 2015 Aug 30;29(16):1521-9. doi: 10.1002/rcm.7244. Rapid Commun Mass Spectrom. 2015. PMID: 26212167
-
Software Tools and Approaches for Compound Identification of LC-MS/MS Data in Metabolomics.Metabolites. 2018 May 10;8(2):31. doi: 10.3390/metabo8020031. Metabolites. 2018. PMID: 29748461 Free PMC article. Review.
-
Searching molecular structure databases using tandem MS data: are we there yet?Curr Opin Chem Biol. 2017 Feb;36:1-6. doi: 10.1016/j.cbpa.2016.12.010. Epub 2016 Dec 22. Curr Opin Chem Biol. 2017. PMID: 28025165 Review.
Cited by
-
Online Prioritization of Toxic Compounds in Water Samples through Intelligent HRMS Data Acquisition.Anal Chem. 2021 Mar 30;93(12):5071-5080. doi: 10.1021/acs.analchem.0c04473. Epub 2021 Mar 16. Anal Chem. 2021. PMID: 33724776 Free PMC article.
-
Navigating common pitfalls in metabolite identification and metabolomics bioinformatics.Metabolomics. 2024 Sep 21;20(5):103. doi: 10.1007/s11306-024-02167-2. Metabolomics. 2024. PMID: 39305388 Free PMC article. Review.
-
Wheat growth, applied water use efficiency and flag leaf metabolome under continuous and pulsed deficit irrigation.Sci Rep. 2020 Jun 22;10(1):10112. doi: 10.1038/s41598-020-66812-1. Sci Rep. 2020. PMID: 32572060 Free PMC article.
-
Food Phenotyping: Recording and Processing of Non-Targeted Liquid Chromatography Mass Spectrometry Data for Verifying Food Authenticity.Molecules. 2020 Aug 31;25(17):3972. doi: 10.3390/molecules25173972. Molecules. 2020. PMID: 32878155 Free PMC article. Review.
-
Estimating LoD-s Based on the Ionization Efficiency Values for the Reporting and Harmonization of Amenable Chemical Space in Nontargeted Screening LC/ESI/HRMS.Anal Chem. 2024 Jul 16;96(28):11263-11272. doi: 10.1021/acs.analchem.4c01002. Epub 2024 Jul 3. Anal Chem. 2024. PMID: 38959408 Free PMC article.
References
-
- Schymanski EL, Singer HP, Slobodnik J, Ipolyi IM, Oswald P, Krauss M, Schulze T, Haglund P, Letzel T, Grosse S, et al. Non-target screening with high-resolution mass spectrometry: critical review using a collaborative trial on water analysis. Anal Bioanal Chem. 2015;407(21):6237–6255. doi: 10.1007/s00216-015-8681-7. - DOI - PubMed
-
- Schymanski EL, Singer HP, Longrée P, Loos M, Ruff M, Stravs MA, Ripollés Vidal C, Hollender J. Strategies to characterize polar organic contamination in wastewater: exploring the capability of high resolution mass spectrometry. Environ Sci Technol. 2014;48(3):1811–1818. doi: 10.1021/es4044374. - DOI - PubMed
-
- Vinaixa M, Schymanski EL, Neumann S, Navarro M, Salek RM, Yanes O (2015) Mass spectral databases for LC/MS and GC/MS-based metabolomics: state of the field and future prospects. Trends Anal Chem (TrAC). doi:10.1016/j.trac.2015.09.005
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

