The ICR1000 UK exome series: a resource of gene variation in an outbred population
- PMID: 26834991
- PMCID: PMC4706061
- DOI: 10.12688/f1000research.7049.1
The ICR1000 UK exome series: a resource of gene variation in an outbred population
Abstract
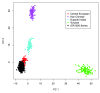
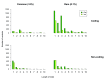
To enhance knowledge of gene variation in outbred populations, and to provide a dataset with utility in research and clinical genomics, we performed exome sequencing of 1,000 UK individuals from the general population and applied a high-quality analysis pipeline that includes high sensitivity and specificity for indel detection. Each UK individual has, on average, 21,978 gene variants including 160 rare (0.1%) variants not present in any other individual in the series. These data provide a baseline expectation for gene variation in an outbred population. Summary data of all 295,391 variants we detected are included here and the individual exome sequences are available from the European Genome-phenome Archive as the ICR1000 UK exome series. Furthermore, samples and other phenotype and experimental data for these individuals are obtainable through application to the 1958 Birth Cohort committee.
Keywords: NGS; exome; exome sequencing; gene variation; next-generation sequencing; population genetics; variant.
Conflict of interest statement
Figures



References
-
- Exome Aggregation Consortium (ExAC).Cambridge, MA. Accessed: December2014. Reference Source
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

