Automatic classification framework for ventricular septal defects: a pilot study on high-throughput mouse embryo cardiac phenotyping
- PMID: 26835488
- PMCID: PMC4717189
- DOI: 10.1117/1.JMI.2.4.041003
Automatic classification framework for ventricular septal defects: a pilot study on high-throughput mouse embryo cardiac phenotyping
Abstract
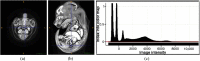
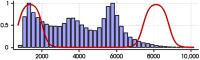
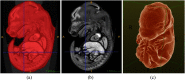
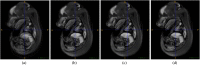
Intensive international efforts are underway toward phenotyping the entire mouse genome by modifying all its [Formula: see text] genes one-by-one for comparative studies. A workload of this scale has triggered numerous studies harnessing image informatics for the identification of morphological defects. However, existing work in this line primarily rests on abnormality detection via structural volumetrics between wild-type and gene-modified mice, which generally fails when the pathology involves no severe volume changes, such as ventricular septal defects (VSDs) in the heart. Furthermore, in embryo cardiac phenotyping, the lack of relevant work in embryonic heart segmentation, the limited availability of public atlases, and the general requirement of manual labor for the actual phenotype classification after abnormality detection, along with other limitations, have collectively restricted existing practices from meeting the high-throughput demands. This study proposes, to the best of our knowledge, the first fully automatic VSD classification framework in mouse embryo imaging. Our approach leverages a combination of atlas-based segmentation and snake evolution techniques to derive the segmentation of heart ventricles, where VSD classification is achieved by checking whether the left and right ventricles border or overlap with each other. A pilot study has validated our approach at a proof-of-concept level and achieved a classification accuracy of 100% through a series of empirical experiments on a database of 15 images.
Keywords: atlas-based segmentation; mouse embryo phenotyping; snake evolution; ventricular septal defects.
Figures



Similar articles
-
Principal component analysis-based features generation combined with ellipse models-based classification criterion for a ventricular septal defect diagnosis system.Australas Phys Eng Sci Med. 2018 Dec;41(4):821-836. doi: 10.1007/s13246-018-0676-1. Epub 2018 Sep 20. Australas Phys Eng Sci Med. 2018. PMID: 30238221
-
Evaluation of ventricular septal defects in horses using two-dimensional and Doppler echocardiography.Equine Vet J Suppl. 1995 Sep;(19):86-95. doi: 10.1111/j.2042-3306.1995.tb04994.x. Equine Vet J Suppl. 1995. PMID: 8933074
-
Image-based clustering and connected component labeling for rapid automated left and right ventricular endocardial volume extraction and segmentation in full cardiac cycle multi-frame MRI images of cardiac patients.Med Biol Eng Comput. 2019 Jun;57(6):1213-1228. doi: 10.1007/s11517-019-01952-9. Epub 2019 Jan 28. Med Biol Eng Comput. 2019. PMID: 30690663
-
Classification of Ventricular Septal Defects for the Eleventh Iteration of the International Classification of Diseases-Striving for Consensus: A Report From the International Society for Nomenclature of Paediatric and Congenital Heart Disease.Ann Thorac Surg. 2018 Nov;106(5):1578-1589. doi: 10.1016/j.athoracsur.2018.06.020. Epub 2018 Jul 19. Ann Thorac Surg. 2018. PMID: 30031844 Review.
-
Recent advances in managing septal defects: ventricular septal defects and atrioventricular septal defects.F1000Res. 2018 Apr 26;7:F1000 Faculty Rev-498. doi: 10.12688/f1000research.14102.1. eCollection 2018. F1000Res. 2018. PMID: 29770201 Free PMC article. Review.
Cited by
-
CACCT: An Automated Tool of Detecting Complicated Cardiac Malformations in Mouse Models.Adv Sci (Weinh). 2020 Feb 20;7(8):1903592. doi: 10.1002/advs.201903592. eCollection 2020 Apr. Adv Sci (Weinh). 2020. PMID: 32328433 Free PMC article.
-
An Overview on Image Registration Techniques for Cardiac Diagnosis and Treatment.Cardiol Res Pract. 2018 Aug 8;2018:1437125. doi: 10.1155/2018/1437125. eCollection 2018. Cardiol Res Pract. 2018. PMID: 30159169 Free PMC article. Review.
References
-
- National Human Genome Research Institute, “An overview of the human genome project,” 2012, http://www.genome.gov/12011238 (19 August 2015).
-
- International Mouse Phenotyping Consortium (IMPC), www.mousephenotype.org

