Genome assembly and geospatial phylogenomics of the bed bug Cimex lectularius
- PMID: 26836631
- PMCID: PMC4740774
- DOI: 10.1038/ncomms10164
Genome assembly and geospatial phylogenomics of the bed bug Cimex lectularius
Abstract
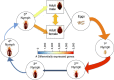
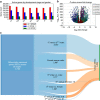
The common bed bug (Cimex lectularius) has been a persistent pest of humans for thousands of years, yet the genetic basis of the bed bug's basic biology and adaptation to dense human environments is largely unknown. Here we report the assembly, annotation and phylogenetic mapping of the 697.9-Mb Cimex lectularius genome, with an N50 of 971 kb, using both long and short read technologies. A RNA-seq time course across all five developmental stages and male and female adults generated 36,985 coding and noncoding gene models. The most pronounced change in gene expression during the life cycle occurs after feeding on human blood and included genes from the Wolbachia endosymbiont, which shows a simultaneous and coordinated host/commensal response to haematophagous activity. These data provide a rich genetic resource for mapping activity and density of C. lectularius across human hosts and cities, which can help track, manage and control bed bug infestations.
Figures


References
-
- Usinger R. Monograph of Cimicidae Vol. 7, (Entomological Society of America, Thomas Say Foundation (1966).
-
- Schuh R. T. & Slater J. A. True Bugs of the World (Hemiptera: Heteroptera): Classification and Natural History Cornell University Press (1995).
-
- Foottit R. G., Adler P. H. & Henry T. J. in Insect Biodiversity: Science and Society 223–263Blackwell Publishing Ltd (2009).
-
- Mellanby K. A comparison of the physiology of the two species of bed-bug which attack man. Parasitology 27, 111–122 (1935).
-
- Potter M. F. The history of bed bug management. Am. Entomol. 57, 14–25 (2011).
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

