Identifying ligands at orphan GPCRs: current status using structure-based approaches
- PMID: 26837045
- PMCID: PMC5341249
- DOI: 10.1111/bph.13452
Identifying ligands at orphan GPCRs: current status using structure-based approaches
Abstract
GPCRs are the most successful pharmaceutical targets in history. Nevertheless, the pharmacology of many GPCRs remains inaccessible as their endogenous or exogenous modulators have not been discovered. Tools that explore the physiological functions and pharmacological potential of these 'orphan' GPCRs, whether they are endogenous and/or surrogate ligands, are therefore of paramount importance. Rates of receptor deorphanization determined by traditional reverse pharmacology methods have slowed, indicating a need for the development of more sophisticated and efficient ligand screening approaches. Here, we discuss the use of structure-based ligand discovery approaches to identify small molecule modulators for exploring the function of orphan GPCRs. These studies have been buoyed by the growing number of GPCR crystal structures solved in the past decade, providing a broad range of template structures for homology modelling of orphans. This review discusses the methods used to establish the appropriate signalling assays to test orphan receptor activity and provides current examples of structure-based methods used to identify ligands of orphan GPCRs. Linked Articles This article is part of a themed section on Molecular Pharmacology of G Protein-Coupled Receptors. To view the other articles in this section visit http://onlinelibrary.wiley.com/doi/10.1111/bph.v173.20/issuetoc.
© 2016 The British Pharmacological Society.
Figures

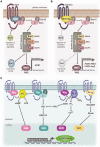
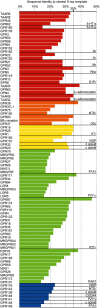
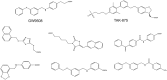
References
-
- Andrews SP, Brown GA, Christopher JA (2014). Structure‐based and fragment‐based GPCR drug discovery. ChemMedChem 9: 256–275. - PubMed
-
- Ballesteros JA, Weinstein H (1995). Integrated methods for the construction of three‐dimensional models and computational probing of structure–function relations in G protein‐coupled receptors In: Sealfon SC. (ed). Receptor Molecular Biology, Methods in Neuroscience, Vol. 25 Academic Press: New York, pp. 366–428.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

