Recurrent chimeric fusion RNAs in non-cancer tissues and cells
- PMID: 26837576
- PMCID: PMC4824105
- DOI: 10.1093/nar/gkw032
Recurrent chimeric fusion RNAs in non-cancer tissues and cells
Abstract
Gene fusions and their products (RNA and protein) were once thought to be unique features to cancer. However, chimeric RNAs can also be found in normal cells. Here, we performed, curated and analyzed nearly 300 RNA-Seq libraries covering 30 different non-neoplastic human tissues and cells as well as 15 mouse tissues. A large number of fusion transcripts were found. Most fusions were detected only once, while 291 were seen in more than one sample. We focused on the recurrent fusions and performed RNA and protein level validations on a subset. We characterized these fusions based on various features of the fusions, and their parental genes. They tend to be expressed at higher levels relative to their parental genes than the non-recurrent ones. Over half of the recurrent fusions involve neighboring genes transcribing in the same direction. A few sequence motifs were found enriched close to the fusion junction sites. We performed functional analyses on a few widely expressed fusions, and found that silencing them resulted in dramatic reduction in normal cell growth and/or motility. Most chimeras use canonical splicing sites, thus are likely products of 'intergenic splicing'. We also explored the implications of these non-pathological fusions in cancer and in evolution.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
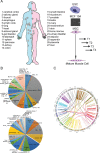

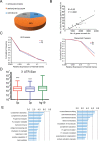

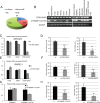
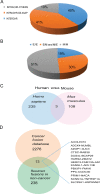
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

