Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia
- PMID: 26837699
- PMCID: PMC4912011
- DOI: 10.1182/blood-2015-07-659144
Clinical impact of clonal and subclonal TP53, SF3B1, BIRC3, NOTCH1, and ATM mutations in chronic lymphocytic leukemia
Abstract
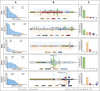
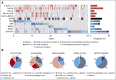
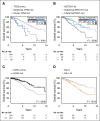
Genomic studies have revealed the complex clonal heterogeneity of chronic lymphocytic leukemia (CLL). The acquisition and selection of genomic aberrations may be critical to understanding the progression of this disease. In this study, we have extensively characterized the mutational status of TP53, SF3B1, BIRC3, NOTCH1, and ATM in 406 untreated CLL cases by ultra-deep next-generation sequencing, which detected subclonal mutations down to 0.3% allele frequency. Clonal dynamics were examined in longitudinal samples of 48 CLL patients. We identified a high proportion of subclonal mutations, isolated or associated with clonal aberrations. TP53 mutations were present in 10.6% of patients (6.4% clonal, 4.2% subclonal), ATM mutations in 11.1% (7.8% clonal, 1.3% subclonal, 2% germ line mutations considered pathogenic), SF3B1 mutations in 12.6% (7.4% clonal, 5.2% subclonal), NOTCH1 mutations in 21.8% (14.2% clonal, 7.6% subclonal), and BIRC3 mutations in 4.2% (2% clonal, 2.2% subclonal). ATM mutations, clonal SF3B1, and both clonal and subclonal NOTCH1 mutations predicted for shorter time to first treatment irrespective of the immunoglobulin heavy-chain variable-region gene (IGHV) mutational status. Clonal and subclonal TP53 and clonal NOTCH1 mutations predicted for shorter overall survival together with the IGHV mutational status. Clonal evolution in longitudinal samples mainly occurred in cases with mutations in the initial samples and was observed not only after chemotherapy but also in untreated patients. These findings suggest that the characterization of the subclonal architecture and its dynamics in the evolution of the disease may be relevant for the management of CLL patients.
© 2016 by The American Society of Hematology.
Figures


Comment in
-
Not all subclones matter in CLL.Blood. 2016 Apr 28;127(17):2052-4. doi: 10.1182/blood-2016-02-699041. Blood. 2016. PMID: 27127300 No abstract available.
References
-
- Zenz T, Mertens D, Küppers R, Döhner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer. 2010;10(1):37–50. - PubMed
-
- Hallek M. Chronic lymphocytic leukemia: 2013 update on diagnosis, risk stratification and treatment. Am J Hematol. 2013;88(9):803–816. - PubMed
-
- Villamor N, López-Guillermo A, López-Otín C, Campo E. Next-generation sequencing in chronic lymphocytic leukemia. Semin Hematol. 2013;50(4):286–295. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

