Major bacterial lineages are essentially devoid of CRISPR-Cas viral defence systems
- PMID: 26837824
- PMCID: PMC4742961
- DOI: 10.1038/ncomms10613
Major bacterial lineages are essentially devoid of CRISPR-Cas viral defence systems
Abstract
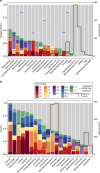
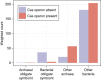
Current understanding of microorganism-virus interactions, which shape the evolution and functioning of Earth's ecosystems, is based primarily on cultivated organisms. Here we investigate thousands of viral and microbial genomes recovered using a cultivation-independent approach to study the frequency, variety and taxonomic distribution of viral defence mechanisms. CRISPR-Cas systems that confer microorganisms with immunity to viruses are present in only 10% of 1,724 sampled microorganisms, compared with previous reports of 40% occurrence in bacteria and 81% in archaea. We attribute this large difference to the lack of CRISPR-Cas systems across major bacterial lineages that have no cultivated representatives. We correlate absence of CRISPR-Cas with lack of nucleotide biosynthesis capacity and a symbiotic lifestyle. Restriction systems are well represented in these lineages and might provide both non-specific viral defence and access to nucleotides.
Figures

References
-
- Barrangou R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007). - PubMed
-
- Garneau J. E. et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67–71 (2010). - PubMed
-
- Sorek R., Kunin V. & Hugenholtz P. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6, 181–186 (2008). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

