Barcode Sequencing Screen Identifies SUB1 as a Regulator of Yeast Pheromone Inducible Genes
- PMID: 26837954
- PMCID: PMC4825658
- DOI: 10.1534/g3.115.026757
Barcode Sequencing Screen Identifies SUB1 as a Regulator of Yeast Pheromone Inducible Genes
Abstract
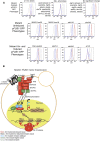
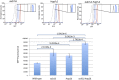
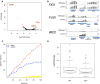
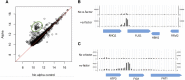
The yeast pheromone response pathway serves as a valuable model of eukaryotic mitogen-activated protein kinase (MAPK) pathways, and transcription of their downstream targets. Here, we describe application of a screening method combining two technologies: fluorescence-activated cell sorting (FACS), and barcode analysis by sequencing (Bar-Seq). Using this screening method, and pFUS1-GFP as a reporter for MAPK pathway activation, we readily identified mutants in known mating pathway components. In this study, we also include a comprehensive analysis of the FUS1 induction properties of known mating pathway mutants by flow cytometry, featuring single cell analysis of each mutant population. We also characterized a new source of false positives resulting from the design of this screen. Additionally, we identified a deletion mutant, sub1Δ, with increased basal expression of pFUS1-GFP. Here, in the first ChIP-Seq of Sub1, our data shows that Sub1 binds to the promoters of about half the genes in the genome (tripling the 991 loci previously reported), including the promoters of several pheromone-inducible genes, some of which show an increase upon pheromone induction. Here, we also present the first RNA-Seq of a sub1Δ mutant; the majority of genes have no change in RNA, but, of the small subset that do, most show decreased expression, consistent with biochemical studies implicating Sub1 as a positive transcriptional regulator. The RNA-Seq data also show that certain pheromone-inducible genes are induced less in the sub1Δ mutant relative to the wild type, supporting a role for Sub1 in regulation of mating pathway genes. The sub1Δ mutant has increased basal levels of a small subset of other genes besides FUS1, including IMD2 and FIG1, a gene encoding an integral membrane protein necessary for efficient mating.
Keywords: SUB1; mating; transcription.
Copyright © 2016 Sliva et al.
Figures


Similar articles
-
MAP Kinase Regulation of the Candida albicans Pheromone Pathway.mSphere. 2019 Feb 20;4(1):e00598-18. doi: 10.1128/mSphere.00598-18. mSphere. 2019. PMID: 30787119 Free PMC article.
-
The yeast MOT2 gene encodes a putative zinc finger protein that serves as a global negative regulator affecting expression of several categories of genes, including mating-pheromone-responsive genes.Mol Cell Biol. 1994 May;14(5):3150-7. doi: 10.1128/mcb.14.5.3150-3157.1994. Mol Cell Biol. 1994. PMID: 8164670 Free PMC article.
-
The Leu-132 of the Ste4(Gbeta) subunit is essential for proper coupling of the G protein with the Ste2 alpha factor receptor during the mating pheromone response in yeast.FEBS Lett. 2000 Feb 4;467(1):22-6. doi: 10.1016/s0014-5793(00)01106-6. FEBS Lett. 2000. PMID: 10664449
-
Basidiomycete mating type genes and pheromone signaling.Eukaryot Cell. 2010 Jun;9(6):847-59. doi: 10.1128/EC.00319-09. Epub 2010 Feb 26. Eukaryot Cell. 2010. PMID: 20190072 Free PMC article. Review.
-
Pheromone signaling pathways in yeast.Sci STKE. 2006 Dec 5;2006(364):cm6. doi: 10.1126/stke.3642006cm6. Sci STKE. 2006. PMID: 17148787 Review.
Cited by
-
Systematic identification of factors mediating accelerated mRNA degradation in response to changes in environmental nitrogen.PLoS Genet. 2018 May 21;14(5):e1007406. doi: 10.1371/journal.pgen.1007406. eCollection 2018 May. PLoS Genet. 2018. PMID: 29782489 Free PMC article.
-
Functional genomics of lipid metabolism in the oleaginous yeast Rhodosporidium toruloides.Elife. 2018 Mar 9;7:e32110. doi: 10.7554/eLife.32110. Elife. 2018. PMID: 29521624 Free PMC article.
-
Genome-Wide Screen for Enhanced Noncanonical Amino Acid Incorporation in Yeast.ACS Synth Biol. 2022 Nov 18;11(11):3669-3680. doi: 10.1021/acssynbio.2c00267. Epub 2022 Nov 8. ACS Synth Biol. 2022. PMID: 36346914 Free PMC article.
References
-
- Bardwell L., 2004. A walk-through of the yeast mating pheromone response pathway. Peptides 25: 1465–1476. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
