Reconstruction of the Evolutionary History and Dispersal of Usutu Virus, a Neglected Emerging Arbovirus in Europe and Africa
- PMID: 26838717
- PMCID: PMC4742707
- DOI: 10.1128/mBio.01938-15
Reconstruction of the Evolutionary History and Dispersal of Usutu Virus, a Neglected Emerging Arbovirus in Europe and Africa
Abstract
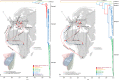
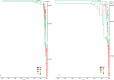
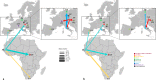
Usutu virus (USUV), one of the most neglected Old World encephalitic flaviviruses, causes epizootics among wild and captive birds and sporadic infection in humans. The dynamics of USUV spread and evolution in its natural hosts are unknown. Here, we present the phylogeny and evolutionary history of all available USUV strains, including 77 newly sequenced complete genomes from a variety of host species at a temporal and spatial scaled resolution. The results showed that USUV can be classified into six distinct lineages and that the most recent common ancestor of the recent European epizootics emerged in Africa at least 500 years ago. We demonstrated that USUV was introduced regularly from Africa into Europe in the last 50 years, and the genetic diversity of European lineages is shaped primarily by in situ evolution, while the African lineages have been driven by extensive gene flow. Most of the amino acid changes are deleterious polymorphisms removed by purifying selection, with adaptive evolution restricted to the NS5 gene and several others evolving under episodic directional selection, indicating that the ecological or immunological factors were mostly the key determinants of USUV dispersal and outbreaks. Host-specific mutations have been detected, while the host transition analysis identified mosquitoes as the most likely origin of the common ancestor and birds as the source of the recent European USUV lineages. Our results suggest that the major migratory bird flyways could predict the continental and intercontinental dispersal patterns of USUV and that migratory birds might act as potential long-distance dispersal vehicles.
Importance: Usutu virus (USUV), a mosquito-borne flavivirus of the Japanese encephalitis virus antigenic group, caused massive bird die-offs, mostly in Europe. There is increasing evidence that USUV appears to be pathogenic for humans, becoming a potential public health problem. The emergence of USUV in Europe allows us to understand how an arbovirus spreads, adapts, and evolves in a naive environment. Thus, understanding the epidemiological and evolutionary processes that contribute to the emergence, maintenance, and further spread of viral diseases is the sine qua non to develop and implement surveillance strategies for their control. In this work, we performed an expansive phylogeographic and evolutionary analysis of USUV using all published sequences and those generated during this study. Subsequently, we described the genetic traits, reconstructed the potential pattern of geographic spread between continents/countries of the identified viral lineages and the drivers of viral migration, and traced the origin of outbreaks and transition events between different hosts.
Copyright © 2016 Engel et al.
Figures


References
-
- Williams MC, Simpson DI, Haddow AJ, Knight EM. 1964. The isolation of West Nile virus from man and of Usutu virus from the bird-biting mosquito Mansonia aurites (Theobald) in the Entebbe area of Uganda. Ann Trop Med Parasitol 58:367–374. - PubMed
-
- McIntosh BM. 1985. Usutu (SAAr 1776), nouvel arbovirus du groupe B. Int Cat Arboviruses 3:1059–1060.
-
- King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed). 2011. Virus taxonomy. Ninth report of the International Committee on Taxonomy of Viruses Elsevier Academic Press, London, United Kingdom.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

