End-to-end automated microfluidic platform for synthetic biology: from design to functional analysis
- PMID: 26839585
- PMCID: PMC4736182
- DOI: 10.1186/s13036-016-0024-5
End-to-end automated microfluidic platform for synthetic biology: from design to functional analysis
Abstract
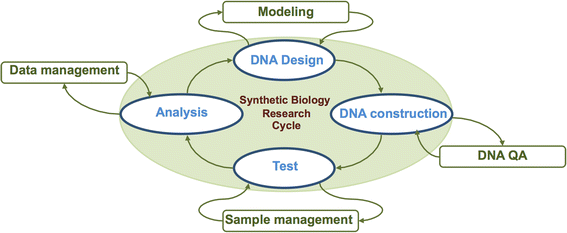
Background: Synthetic biology aims to engineer biological systems for desired behaviors. The construction of these systems can be complex, often requiring genetic reprogramming, extensive de novo DNA synthesis, and functional screening.
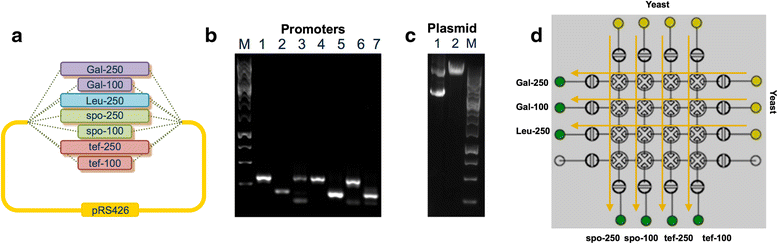
Results: Herein, we present a programmable, multipurpose microfluidic platform and associated software and apply the platform to major steps of the synthetic biology research cycle: design, construction, testing, and analysis. We show the platform's capabilities for multiple automated DNA assembly methods, including a new method for Isothermal Hierarchical DNA Construction, and for Escherichia coli and Saccharomyces cerevisiae transformation. The platform enables the automated control of cellular growth, gene expression induction, and proteogenic and metabolic output analysis.
Conclusions: Taken together, we demonstrate the microfluidic platform's potential to provide end-to-end solutions for synthetic biology research, from design to functional analysis.
Keywords: Analysis; Cell culture; DNA assembly; Microfluidics; Synthetic biology; Transformation.
Figures




Similar articles
-
A Versatile Microfluidic Device for Automating Synthetic Biology.ACS Synth Biol. 2015 Oct 16;4(10):1151-64. doi: 10.1021/acssynbio.5b00062. Epub 2015 Jun 15. ACS Synth Biol. 2015. PMID: 26075958
-
A Droplet Microfluidic Platform for Automating Genetic Engineering.ACS Synth Biol. 2016 May 20;5(5):426-33. doi: 10.1021/acssynbio.6b00011. Epub 2016 Feb 22. ACS Synth Biol. 2016. PMID: 26830031
-
The fusion of biology, computer science, and engineering: towards efficient and successful synthetic biology.Perspect Biol Med. 2012;55(4):503-20. doi: 10.1353/pbm.2012.0044. Perspect Biol Med. 2012. PMID: 23502561
-
Droplet microfluidics for synthetic biology.Lab Chip. 2017 Oct 11;17(20):3388-3400. doi: 10.1039/c7lc00576h. Lab Chip. 2017. PMID: 28820204 Review.
-
Integration of microfluidics into the synthetic biology design flow.Lab Chip. 2014 Sep 21;14(18):3459-74. doi: 10.1039/c4lc00509k. Epub 2014 Jul 11. Lab Chip. 2014. PMID: 25012162 Review.
Cited by
-
Improving engineered biological systems with electronics and microfluidics.Nat Biotechnol. 2025 Jul;43(7):1067-1083. doi: 10.1038/s41587-025-02709-6. Epub 2025 Jun 27. Nat Biotechnol. 2025. PMID: 40579601 Review.
-
The Oxygen Release Instrument: Space Mission Reactive Oxygen Species Measurements for Habitability Characterization, Biosignature Preservation Potential Assessment, and Evaluation of Human Health Hazards.Life (Basel). 2019 Aug 27;9(3):70. doi: 10.3390/life9030070. Life (Basel). 2019. PMID: 31461989 Free PMC article.
-
Hardware, Software, and Wetware Codesign Environment for Synthetic Biology.Biodes Res. 2022 Sep 1;2022:9794510. doi: 10.34133/2022/9794510. eCollection 2022. Biodes Res. 2022. PMID: 37850136 Free PMC article. Review.
-
Process Variability in Top-Down Fabrication of Silicon Nanowire-Based Biosensor Arrays.Sensors (Basel). 2021 Jul 29;21(15):5153. doi: 10.3390/s21155153. Sensors (Basel). 2021. PMID: 34372390 Free PMC article. Review.
-
A pneumatic random-access memory for controlling soft robots.PLoS One. 2021 Jul 16;16(7):e0254524. doi: 10.1371/journal.pone.0254524. eCollection 2021. PLoS One. 2021. PMID: 34270580 Free PMC article.
References
-
- Rollie S, Mangold M, Sundmacher K. Designing biological systems: systems engineering meets synthetic biology. Chem Eng Sci. 2012;69(1):1–29. doi: 10.1016/j.ces.2011.10.068. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

