One-step multiplex real-time RT-PCR assay for detecting and genotyping wild-type group A rotavirus strains and vaccine strains (Rotarix® and RotaTeq®) in stool samples
- PMID: 26839745
- PMCID: PMC4734446
- DOI: 10.7717/peerj.1560
One-step multiplex real-time RT-PCR assay for detecting and genotyping wild-type group A rotavirus strains and vaccine strains (Rotarix® and RotaTeq®) in stool samples
Abstract
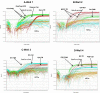
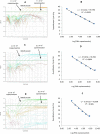
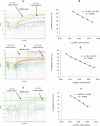
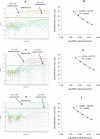
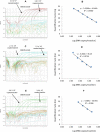
Background. Group A rotavirus (RVA) infection is the major cause of acute gastroenteritis (AGE) in young children worldwide. Introduction of two live-attenuated rotavirus vaccines, RotaTeq® and Rotarix®, has dramatically reduced RVA associated AGE and mortality in developed as well as in many developing countries. High-throughput methods are needed to genotype rotavirus wild-type strains and to identify vaccine strains in stool samples. Quantitative RT-PCR assays (qRT-PCR) offer several advantages including increased sensitivity, higher throughput, and faster turnaround time. Methods. In this study, a one-step multiplex qRT-PCR assay was developed to detect and genotype wild-type strains and vaccine (Rotarix® and RotaTeq®) rotavirus strains along with an internal processing control (Xeno or MS2 RNA). Real-time RT-PCR assays were designed for VP7 (G1, G2, G3, G4, G9, G12) and VP4 (P[4], P[6] and P[8]) genotypes. The multiplex qRT-PCR assay also included previously published NSP3 qRT-PCR for rotavirus detection and Rotarix® NSP2 and RotaTeq® VP6 qRT-PCRs for detection of Rotarix® and RotaTeq® vaccine strains respectively. The multiplex qRT-PCR assay was validated using 853 sequence confirmed stool samples and 24 lab cultured strains of different rotavirus genotypes. By using thermostable rTth polymerase enzyme, dsRNA denaturation, reverse transcription (RT) and amplification (PCR) steps were performed in single tube by uninterrupted thermocycling profile to reduce chances of sample cross contamination and for rapid generation of results. For quantification, standard curves were generated using dsRNA transcripts derived from RVA gene segments. Results. The VP7 qRT-PCRs exhibited 98.8-100% sensitivity, 99.7-100% specificity, 85-95% efficiency and a limit of detection of 4-60 copies per singleplex reaction. The VP7 qRT-PCRs exhibited 81-92% efficiency and limit of detection of 150-600 copies in multiplex reactions. The VP4 qRT-PCRs exhibited 98.8-100% sensitivity, 100% specificity, 86-89% efficiency and a limit of detection of 12-400 copies per singleplex reactions. The VP4 qRT-PCRs exhibited 82-90% efficiency and limit of detection of 120-4000 copies in multiplex reaction. Discussion. The one-step multiplex qRT-PCR assay will facilitate high-throughput rotavirus genotype characterization for monitoring circulating rotavirus wild-type strains causing rotavirus infections, determining the frequency of Rotarix® and RotaTeq® vaccine strains and vaccine-derived reassortants associated with AGE, and help to identify novel rotavirus strains derived by reassortment between vaccine and wild-type strains.
Keywords: Gastroenteritis; Multiplex qRT-PCR; RotaTeq®; Rotarix®; Rotavirus genotyping.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
-
- Banyai K, Laszlo B, Duque J, Steele AD, Nelson EA, Gentsch JR, Parashar UD. Systematic review of regional and temporal trends in global rotavirus strain diversity in the pre rotavirus vaccine era: insights for understanding the impact of rotavirus vaccination programs. Vaccine. 2012;30(Suppl 1):A122–130. doi: 10.1016/j.vaccine.2011.09.111. - DOI - PubMed
-
- Boom JA, Sahni LC, Payne DC, Gautam R, Lyde F, Mijatovic-Rustempasic S, Bowen MD, Tate JE, Rench MA, Gentsch JR, Parashar UD, Baker CJ. Symptomatic infection and detection of vaccine and vaccine-reassortant rotavirus strains in 5 children: a case series. Journal of Infectious Diseases. 2012;206(8):1275–1279. doi: 10.1093/infdis/jis490. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

