Small molecule screen for inhibitors of expression from canonical CREB response element-containing promoters
- PMID: 26840025
- PMCID: PMC4890994
- DOI: 10.18632/oncotarget.7085
Small molecule screen for inhibitors of expression from canonical CREB response element-containing promoters
Abstract
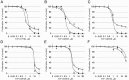
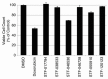
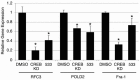
The transcription factor CREB (cAMP Response Element Binding Protein) is an important determinant in the growth of Acute Myeloid Leukemia (AML) cells. CREB overexpression increases AML cell growth by driving the expression of key regulators of apoptosis and the cell cycle. Conversely, CREB knockdown inhibits proliferation and survival of AML cells but not normal hematopoietic cells. Thus, CREB represents a promising drug target for the treatment of AML, which carries a poor prognosis. In this study, we performed a high-throughput small molecule screen to identify compounds that disrupt CREB function in AML cells. We screened ~114,000 candidate compounds from Stanford University's small molecule library, and identified 5 molecules that inhibit CREB function at micromolar concentrations, but are non-toxic to normal hematopoietic cells. This study suggests that targeting CREB function using small molecules could provide alternative approaches to treat AML.
Keywords: CREB; novel therapeutics; small molecule screen.
Conflict of interest statement
The authors have declared no financial or other conflicts of interest.
Figures



References
-
- Kornblau SM, Hu CW, Qiu Y, Yoo SY, Chauhan K, Jabbour E, Ravandi F, Coombes K, Qutub AA. CREB/ATF Family Protein Expression States in AML: Active CREB1, but not ATF is an Adverse Prognostic Factor. American Society of Hematology Annual Meeting; San Francisco, CA. 2014.
-
- Shankar DB, Cheng JC, Sakamoto KM. Role of cyclic AMP response element binding protein in human leukemias. Cancer. 2005;104:1819–1824. - PubMed
-
- Shankar DB, Sakamoto KM. The role of cyclic-AMP binding protein (CREB) in leukemia cell proliferation and acute leukemias. Leukemia & lymphoma. 2004;45:265–270. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

