Amyloid precursor protein and amyloid precursor-like protein 2 in cancer
- PMID: 26840089
- PMCID: PMC4991393
- DOI: 10.18632/oncotarget.7103
Amyloid precursor protein and amyloid precursor-like protein 2 in cancer
Abstract
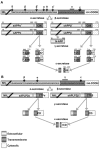
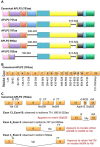
Amyloid precursor protein (APP) and its family members amyloid precursor-like protein 1 (APLP1) and amyloid precursor-like protein 2 (APLP2) are type 1 transmembrane glycoproteins that are highly conserved across species. The transcriptional regulation of APP and APLP2 is similar but not identical, and the cleavage of both proteins is regulated by phosphorylation. APP has been implicated in Alzheimer's disease causation, and in addition to its importance in neurology, APP is deregulated in cancer cells. APLP2 is likewise overexpressed in cancer cells, and APLP2 and APP are linked to increased tumor cell proliferation, migration, and invasion. In this present review, we discuss the unfolding account of these APP family members' roles in cancer progression and metastasis.
Keywords: amyloid precursor protein; amyloid precursor-like protein 2; cancer; growth; migration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Sprecher CA, Grant FJ, Grimm G, O'Hara PJ, Norris F, Norris K, Foster DC. Molecular cloning of the cDNA for a human amyloid precursor protein homolog: evidence for a multigene family. Biochemistry. 1993;32:4481–4486. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

