Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival
- PMID: 26840267
- PMCID: PMC4891103
- DOI: 10.18632/oncotarget.7067
Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival
Abstract
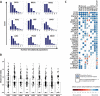
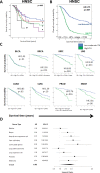
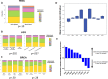
As tumors accumulate genetic alterations, an evolutionary process occurs in which genetically distinct subclonal populations of cells co-exist, resulting in intratumor genetic heterogeneity (ITH). The clinical implications of ITH remain poorly defined. Data are limited with respect to whether ITH is an independent determinant of patient survival outcomes, across different cancer types. Here, we report the results of a pan-cancer analysis of over 3300 tumors, showing a varied landscape of ITH across 9 cancer types. While some gene mutations are subclonal, the majority of driver gene mutations are clonal events, present in nearly all cancer cells. Strikingly, high levels of ITH are associated with poorer survival across diverse types of cancer. The adverse impact of high ITH is independent of other clinical, pathologic and molecular factors. High ITH tends to be associated with lower levels of tumor-infiltrating immune cells, but this association is not able to explain the observed survival differences. Together, these data show that ITH is a prognostic marker in multiple cancers. These results illuminate the natural history of cancer evolution, indicating that tumor heterogeneity represents a significant obstacle to cancer control.
Keywords: cancer; evolution; heterogeneity; immune surveillance; survival.
Conflict of interest statement
None.
Figures
References
-
- Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. - PubMed
-
- de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, Jamal-Hanjani M, Shafi S, Murugaesu N, Rowan AJ, Gronroos E, Muhammad MA, Horswell S, Gerlinger M, Varela I, Jones D, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. - PMC - PubMed
-
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. The New England journal of medicine. 2012;366:883–892. - PMC - PubMed
-
- Yates LR, Gerstung M, Knappskog S, Desmedt C, Gundem G, Van Loo P, Aas T, Alexandrov LB, Larsimont D, Davies H, Li Y, Ju YS, Ramakrishna M, Haugland HK, Lilleng PK, Nik-Zainal S, et al. Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nature medicine. 2015;21:751–759. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

