Germline polymorphisms in an enhancer of PSIP1 are associated with progression-free survival in epithelial ovarian cancer
- PMID: 26840454
- PMCID: PMC4872719
- DOI: 10.18632/oncotarget.7047
Germline polymorphisms in an enhancer of PSIP1 are associated with progression-free survival in epithelial ovarian cancer
Abstract
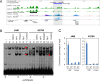
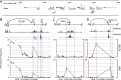
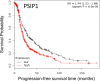
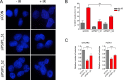
Women with epithelial ovarian cancer (EOC) are usually treated with platinum/taxane therapy after cytoreductive surgery but there is considerable inter-individual variation in response. To identify germline single-nucleotide polymorphisms (SNPs) that contribute to variations in individual responses to chemotherapy, we carried out a multi-phase genome-wide association study (GWAS) in 1,244 women diagnosed with serous EOC who were treated with the same first-line chemotherapy, carboplatin and paclitaxel. We identified two SNPs (rs7874043 and rs72700653) in TTC39B (best P=7x10-5, HR=1.90, for rs7874043) associated with progression-free survival (PFS). Functional analyses show that both SNPs lie in a putative regulatory element (PRE) that physically interacts with the promoters of PSIP1, CCDC171 and an alternative promoter of TTC39B. The C allele of rs7874043 is associated with poor PFS and showed increased binding of the Sp1 transcription factor, which is critical for chromatin interactions with PSIP1. Silencing of PSIP1 significantly impaired DNA damage-induced Rad51 nuclear foci and reduced cell viability in ovarian cancer lines. PSIP1 (PC4 and SFRS1 Interacting Protein 1) is known to protect cells from stress-induced apoptosis, and high expression is associated with poor PFS in EOC patients. We therefore suggest that the minor allele of rs7874043 confers poor PFS by increasing PSIP1 expression.
Keywords: PSIP1; chromosome conformation capture; epithelial ovarian cancer; genome-wide association study; progression free survival.
Conflict of interest statement
The authors disclose no potential conflicts of interest.
Figures



References
-
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. - PubMed
-
- Markman M. Pharmaceutical management of ovarian cancer : current status. Drugs. 2008;68:771–89. - PubMed
-
- Marchetti C, Pisano C, Facchini G, Bruni GS, Magazzino FP, Losito S, Pignata S. First-line treatment of advanced ovarian cancer: current research and perspectives. Expert Rev Anticancer Ther. 2010;10:47–60. - PubMed
-
- Johnatty SE, Beesley J, Paul J, Fereday S, Spurdle AB, Webb PM, Byth K, Marsh S, McLeod H, Group AS, Harnett PR, Brown R, DeFazio A, et al. ABCB1 (MDR 1) polymorphisms and progression-free survival among women with ovarian cancer following paclitaxel/carboplatin chemotherapy. Clin Cancer Res. 2008;14:5594–601. - PubMed
-
- Tian C, Ambrosone CB, Darcy KM, Krivak TC, Armstrong DK, Bookman MA, Davis W, Zhao H, Moysich K, Gallion H, DeLoia JA. Common variants in ABCB1, ABCC2 and ABCG2 genes and clinical outcomes among women with advanced stage ovarian cancer treated with platinum and taxane-based chemotherapy: a Gynecologic Oncology Group study. Gynecol Oncol. 2012;124:575–81. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 GM061393/GM/NIGMS NIH HHS/United States
- P30 CA014599/CA/NCI NIH HHS/United States
- K08 GM089941/GM/NIGMS NIH HHS/United States
- R21 CA139278/CA/NCI NIH HHS/United States
- K08GM089941/GM/NIGMS NIH HHS/United States
- P50 CA136393/CA/NCI NIH HHS/United States
- R01 CA114343/CA/NCI NIH HHS/United States
- C536/A6689/CRUK_/Cancer Research UK/United Kingdom
- P30 CA14599/CA/NCI NIH HHS/United States
- UO1GM61393/GM/NIGMS NIH HHS/United States
- R01 CA122443/CA/NCI NIH HHS/United States
- P30 CA015083/CA/NCI NIH HHS/United States
- R01 CA 61107/CA/NCI NIH HHS/United States
- 13086/CRUK_/Cancer Research UK/United Kingdom
- 16561/CRUK_/Cancer Research UK/United Kingdom
- 10124/CRUK_/Cancer Research UK/United Kingdom
- R01CA114343/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

