Role of Electroosmosis in the Permeation of Neutral Molecules: CymA and Cyclodextrin as an Example
- PMID: 26840725
- PMCID: PMC4744173
- DOI: 10.1016/j.bpj.2015.12.027
Role of Electroosmosis in the Permeation of Neutral Molecules: CymA and Cyclodextrin as an Example
Abstract
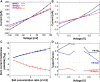
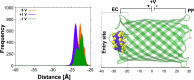
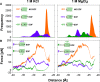
To quantify the flow of small uncharged molecules into and across nanopores, one often uses ion currents. The respective ion-current fluctuations caused by the presence of the analyte make it possible to draw some conclusions about the direction and magnitude of the analyte flow. However, often this flow appears to be asymmetric with respect to the applied voltage. As a possible reason for this asymmetry, we identified the electroosmotic flow (EOF), which is the water transport associated with ions driven by the external transmembrane voltage. As an example, we quantify the contribution of the EOF through a nanopore by investigating the permeation of α-cyclodextrin through CymA, a cyclodextrin-specific channel from Klebsiella oxytoca. To understand the results from electrophysiology on a molecular level, all-atom molecular dynamics simulations are used to detail the effect of the EOF on substrate entry to and exit from a CymA channel in which the N-terminus has been deleted. The combined experimental and computational results strongly suggest that one needs to account for the significant contribution of the EOF when analyzing the penetration of cyclodextrins through the CymA pore. This example study at the same time points to the more general finding that the EOF needs to be considered in translocation studies of neutral molecules and, at least in many cases, should be able to help in discriminating between translocation and binding events.
Copyright © 2016 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures




Similar articles
-
Large-Peptide Permeation Through a Membrane Channel: Understanding Protamine Translocation Through CymA from Klebsiella Oxytoca*.Angew Chem Int Ed Engl. 2021 Apr 6;60(15):8089-8094. doi: 10.1002/anie.202016943. Epub 2021 Mar 3. Angew Chem Int Ed Engl. 2021. PMID: 33580541 Free PMC article.
-
Changes in Salt Concentration Modify the Translocation of Neutral Molecules through a ΔCymA Nanopore in a Non-monotonic Manner.ACS Nano. 2022 May 24;16(5):7701-7712. doi: 10.1021/acsnano.1c11471. Epub 2022 Apr 18. ACS Nano. 2022. PMID: 35435659
-
Outer-membrane translocation of bulky small molecules by passive diffusion.Proc Natl Acad Sci U S A. 2015 Jun 9;112(23):E2991-9. doi: 10.1073/pnas.1424835112. Epub 2015 May 26. Proc Natl Acad Sci U S A. 2015. PMID: 26015567 Free PMC article.
-
Simulations of outer membrane channels and their permeability.Biochim Biophys Acta. 2016 Jul;1858(7 Pt B):1760-71. doi: 10.1016/j.bbamem.2015.12.020. Epub 2015 Dec 23. Biochim Biophys Acta. 2016. PMID: 26721326 Review.
-
Drug permeation through biomembranes: cyclodextrins and the unstirred water layer.Pharmazie. 2012 May;67(5):363-70. Pharmazie. 2012. PMID: 22764564 Review.
Cited by
-
Identification and Functional Characterization of a Novel OprD-like Chitin Uptake Channel in Non-chitinolytic Bacteria.J Biol Chem. 2016 Jun 24;291(26):13622-33. doi: 10.1074/jbc.M116.728881. Epub 2016 May 12. J Biol Chem. 2016. PMID: 27226611 Free PMC article.
-
An antibiotic-resistance conferring mutation in a neisserial porin: Structure, ion flux, and ampicillin binding.Biochim Biophys Acta Biomembr. 2021 Jun 1;1863(6):183601. doi: 10.1016/j.bbamem.2021.183601. Epub 2021 Mar 3. Biochim Biophys Acta Biomembr. 2021. PMID: 33675718 Free PMC article.
-
Conformational flexibility driving charge-selective substrate translocation across a bacterial transporter.Chem Sci. 2024 May 13;15(24):9333-9344. doi: 10.1039/d4sc00345d. eCollection 2024 Jun 19. Chem Sci. 2024. PMID: 38903220 Free PMC article.
-
Large-Peptide Permeation Through a Membrane Channel: Understanding Protamine Translocation Through CymA from Klebsiella Oxytoca*.Angew Chem Int Ed Engl. 2021 Apr 6;60(15):8089-8094. doi: 10.1002/anie.202016943. Epub 2021 Mar 3. Angew Chem Int Ed Engl. 2021. PMID: 33580541 Free PMC article.
-
Recent Advances in Modeling Membrane β-Barrel Proteins Using Molecular Dynamics Simulations: From Their Lipid Environments to Their Assemblies.Methods Mol Biol. 2024;2778:311-330. doi: 10.1007/978-1-0716-3734-0_19. Methods Mol Biol. 2024. PMID: 38478286
References
-
- Singh P.R., Bárcena-Uribarri I., Mahendran K.R. Pulling peptides across nanochannels: resolving peptide binding and translocation through the hetero-oligomeric channel from Nocardia farcinica. ACS Nano. 2012;6:10699–10707. - PubMed
-
- Bezrukov S.M., Kullman L., Winterhalter M. Probing sugar translocation through maltoporin at the single channel level. FEBS Lett. 2000;476:224–228. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

