Replication stress: getting back on track
- PMID: 26840898
- PMCID: PMC5125612
- DOI: 10.1038/nsmb.3163
Replication stress: getting back on track
Abstract
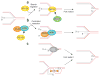
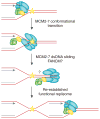
The replication-stress response enables the DNA replication machinery to overcome DNA lesions or intrinsic replication-fork obstacles, and it is essential to ensure faithful transmission of genetic information to daughter cells. Multiple replication stress–response pathways have been identified in recent years, thus raising questions about the specific and possibly redundant functions of these pathways. Here, we review the emerging mechanisms of the replication-stress response in mammalian cells and consider how they may influence the dynamics of the core DNA replication complex.
Figures

References
-
- Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–7. - PubMed
-
- De Piccoli G, et al. Replisome stability at defective DNA replication forks is independent of S phase checkpoint kinases. Mol Cell. 2012;45:696–704. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

