iTRAQ-Based Quantitative Proteomic Analysis of Cotton Roots and Leaves Reveals Pathways Associated with Salt Stress
- PMID: 26841024
- PMCID: PMC4739606
- DOI: 10.1371/journal.pone.0148487
iTRAQ-Based Quantitative Proteomic Analysis of Cotton Roots and Leaves Reveals Pathways Associated with Salt Stress
Abstract
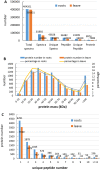
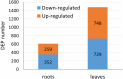
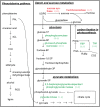
Salinity is a major abiotic stress that affects plant growth and development. In this study, we performed a proteomic analysis of cotton roots and leaf tissue following exposure to saline stress. 611 and 1477 proteins were differentially expressed in the roots and leaves, respectively. In the roots, 259 (42%) proteins were up-regulated and 352 (58%) were down-regulated. In the leaves, 748 (51%) proteins were up-regulated and 729 (49%) were down-regulated. On the basis of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, we concluded that the phenylalanine metabolism and starch and sucrose metabolism were active for energy homeostasis to cope with salt stress in cotton roots. Moreover, photosynthesis, pyruvate metabolism, glycolysis / gluconeogenesis, carbon fixation in photosynthetic organisms and phenylalanine metabolism were inhabited to reduce energy consumption. Characterization of the signaling pathways will help elucidate the mechanism activated by cotton in response to salt stress.
Conflict of interest statement
Figures

References
-
- Nohzadeh Malakshah S, Habibi Rezaei M, Heidari M, Hosseini Salekdeh G. Proteomics reveals new salt responsive proteins associated with rice plasma membrane. Bioscience, Biotechnology, and Biochemistry. 2007; 71: 2144–2154. - PubMed
-
- Munns R. Genes and salt tolerance: bringing them together. New Phytologist. 2005; 167: 645–663. - PubMed
-
- Greenway H, Munns R. Mechanisnn of salt tolerance in non-halophytes. Annu Rev Plant Physiology. 1980: 149–190.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

