Biochemometrics for Natural Products Research: Comparison of Data Analysis Approaches and Application to Identification of Bioactive Compounds
- PMID: 26841051
- PMCID: PMC5135737
- DOI: 10.1021/acs.jnatprod.5b01014
Biochemometrics for Natural Products Research: Comparison of Data Analysis Approaches and Application to Identification of Bioactive Compounds
Abstract
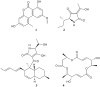
A central challenge of natural products research is assigning bioactive compounds from complex mixtures. The gold standard approach to address this challenge, bioassay-guided fractionation, is often biased toward abundant, rather than bioactive, mixture components. This study evaluated the combination of bioassay-guided fractionation with untargeted metabolite profiling to improve active component identification early in the fractionation process. Key to this methodology was statistical modeling of the integrated biological and chemical data sets (biochemometric analysis). Three data analysis approaches for biochemometric analysis were compared, namely, partial least-squares loading vectors, S-plots, and the selectivity ratio. Extracts from the endophytic fungi Alternaria sp. and Pyrenochaeta sp. with antimicrobial activity against Staphylococcus aureus served as test cases. Biochemometric analysis incorporating the selectivity ratio performed best in identifying bioactive ions from these extracts early in the fractionation process, yielding altersetin (3, MIC 0.23 μg/mL) and macrosphelide A (4, MIC 75 μg/mL) as antibacterial constituents from Alternaria sp. and Pyrenochaeta sp., respectively. This study demonstrates the potential of biochemometrics coupled with bioassay-guided fractionation to identify bioactive mixture components. A benefit of this approach is the ability to integrate multiple stages of fractionation and bioassay data into a single analysis.
Figures




References
-
- Kinghorn AD, Fong HHS, Farnsworth NR, Mehta RG, Moon RC, Moriarty RM, Pezzuto JM. Curr. Org. Chem. 1998;2:597–612.
-
- Wall ME, Wani MC. J. Ethnopharm. 1996;51:239–253. - PubMed
-
- Oberlies NH, Kroll DJ. J. Nat. Prod. 2004;67:129–135. - PubMed
-
- Tu Y. Nat. Med. 2011;17:1217–1220. - PubMed
-
- Noble RL. Biochem. Cell Biol. 1990;68:1344–1351. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

