Transcriptome and Expression Patterns of Chemosensory Genes in Antennae of the Parasitoid Wasp Chouioia cunea
- PMID: 26841106
- PMCID: PMC4739689
- DOI: 10.1371/journal.pone.0148159
Transcriptome and Expression Patterns of Chemosensory Genes in Antennae of the Parasitoid Wasp Chouioia cunea
Abstract
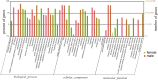
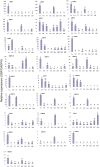
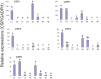
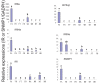
Chouioia cunea Yang is an endoparasitic wasp that attacks pupae of Hyphantria cunea (Drury), an invasive moth species that severely damages forests in China. Chemosensory systems of insects are used to detect volatile chemical odors such as female sex pheromones and host plant volatiles. The antennae of parasite wasps are important for host detection and other sensory-mediated behaviors. We identified and documented differential expression profiles of chemoreception genes in C. cunea antennae. A total of 25 OBPs, 80 ORs, 10 IRs, 11 CSP, 1 SNMPs, and 17 GRs were annotated from adult male and female C. cunea antennal transcriptomes. The expression profiles of 25 OBPs, 16 ORs, and 17 GRs, 5 CSP, 5 IRs and 1 SNMP were determined by RT-PCR and RT-qPCR for the antenna, head, thorax, and abdomen of male and female C. cunea. A total of 8 OBPs, 14 ORs, and 8 GRs, 1 CSP, 4 IRs and 1 SNMPs were exclusively or primarily expressed in female antennae. These female antennal-specific or dominant expression profiles may assist in locating suitable host and oviposition sites. These genes will provide useful targets for advanced study of their biological functions.
Conflict of interest statement
Figures







Similar articles
-
Analysis of the Antennal Transcriptome and Insights into Olfactory Genes in Hyphantria cunea (Drury).PLoS One. 2016 Oct 14;11(10):e0164729. doi: 10.1371/journal.pone.0164729. eCollection 2016. PLoS One. 2016. PMID: 27741298 Free PMC article.
-
Sex Differences in Antennal Transcriptome of Hyphantria cunea and Analysis of Odorant Receptor Expression Profiles.Int J Mol Sci. 2024 Aug 21;25(16):9070. doi: 10.3390/ijms25169070. Int J Mol Sci. 2024. PMID: 39201754 Free PMC article.
-
Identification and sex-biased profiles of candidate olfactory genes in the antennal transcriptome of the parasitoid wasp Cotesia vestalis.Comp Biochem Physiol Part D Genomics Proteomics. 2020 Jun;34:100657. doi: 10.1016/j.cbd.2020.100657. Epub 2020 Jan 29. Comp Biochem Physiol Part D Genomics Proteomics. 2020. PMID: 32032903
-
Antennal transcriptome analysis and comparison of chemosensory gene families in two closely related noctuidae moths, Helicoverpa armigera and H. assulta.PLoS One. 2015 Feb 6;10(2):e0117054. doi: 10.1371/journal.pone.0117054. eCollection 2015. PLoS One. 2015. PMID: 25659090 Free PMC article.
-
Molecular characterization and differential expression of olfactory genes in the antennae of the black cutworm moth Agrotis ipsilon.PLoS One. 2014 Aug 1;9(8):e103420. doi: 10.1371/journal.pone.0103420. eCollection 2014. PLoS One. 2014. PMID: 25083706 Free PMC article.
Cited by
-
Identification and expression profile of odorant-binding proteins in the parasitic wasp Microplitis pallidipes using PacBio long-read sequencing.Parasite. 2022;29:53. doi: 10.1051/parasite/2022053. Epub 2022 Nov 9. Parasite. 2022. PMID: 36350195 Free PMC article.
-
Next-generation biological control: the need for integrating genetics and genomics.Biol Rev Camb Philos Soc. 2020 Dec;95(6):1838-1854. doi: 10.1111/brv.12641. Epub 2020 Aug 14. Biol Rev Camb Philos Soc. 2020. PMID: 32794644 Free PMC article. Review.
-
Insights Into Chemosensory Proteins From Non-Model Insects: Advances and Perspectives in the Context of Pest Management.Front Physiol. 2022 Aug 22;13:924750. doi: 10.3389/fphys.2022.924750. eCollection 2022. Front Physiol. 2022. PMID: 36072856 Free PMC article. Review.
-
Identification and characterization of soluble binding proteins associated with host foraging in the parasitoid wasp Diachasmimorpha longicaudata.PLoS One. 2021 Jun 17;16(6):e0252765. doi: 10.1371/journal.pone.0252765. eCollection 2021. PLoS One. 2021. PMID: 34138896 Free PMC article.
-
Has gene expression neofunctionalization in the fire ant antennae contributed to queen discrimination behavior?Ecol Evol. 2019 Oct 29;9(22):12754-12766. doi: 10.1002/ece3.5748. eCollection 2019 Nov. Ecol Evol. 2019. PMID: 31788211 Free PMC article.
References
-
- Schoonhoven LM, van Loon JJA, Dicke M. Insect-plant biology. 2nd ed New York: Oxford University Press; 2005.
-
- Barbarossa IT, Muroni P, Dardani M, Casula P, Angioy AM. New insight into the antennal chemosensory function of Opius concolor (Hymenoptera, Braconidae). Ital J Zool. 1998; 65: 367–70.
-
- Jacquin-Joly E, Merlin C. Insect olfactory receptors: contributions of molecular biology to chemical ecology. J Chem Ecol. 2004; 30(12): 2359–2397. - PubMed
-
- Hallem E, Ho M, Carlson JR. The molecular basis of odor coding in the Drosophila antenna. Cell. 2004; 117(7): 965–79. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

