Cross Reactive Material 197 glycoconjugate vaccines contain privileged conjugation sites
- PMID: 26841683
- PMCID: PMC4740906
- DOI: 10.1038/srep20488
Cross Reactive Material 197 glycoconjugate vaccines contain privileged conjugation sites
Abstract
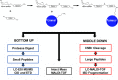
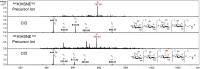
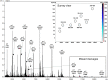
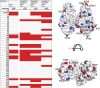
Production of glycoconjugate vaccines involves the chemical conjugation of glycans to an immunogenic carrier protein such as Cross-Reactive-Material-197 (CRM197). Instead of using glycans from natural sources recent vaccine development has been focusing on the use of synthetically defined minimal epitopes. While the glycan is structurally defined, the attachment sites on the protein are not. Fully characterized conjugates and batch-to-batch comparisons are the key to eventually create completely defined conjugates. A variety of glycoconjugates consisting of CRM197 and synthetic oligosaccharide epitopes was characterised using mass spectrometry techniques. The primary structure was assessed by combining intact protein MALDI-TOF-MS, LC-MALDI-TOF-MS middle-down and LC-ESI-MS bottom-up approaches. The middle-down approach on CNBr cleaved glycopeptides provided almost complete sequence coverage, facilitating rapid batch-to-batch comparisons, resolving glycan loading and identification of side products. Regions close to the N- and C-termini were most efficiently conjugated.
Conflict of interest statement
A.R and D.S. are employees of Bruker,Bremen.The other authors declare no competing financial interests.
Figures




Similar articles
-
Determination of glycation sites by tandem mass spectrometry in a synthetic lactose-bovine serum albumin conjugate, a vaccine model prepared by dialkyl squarate chemistry.Rapid Commun Mass Spectrom. 2012 Apr 15;26(7):749-58. doi: 10.1002/rcm.6166. Rapid Commun Mass Spectrom. 2012. PMID: 22368054 Free PMC article.
-
Carrier priming with CRM 197 or diphtheria toxoid has a different impact on the immunogenicity of the respective glycoconjugates: biophysical and immunochemical interpretation.Vaccine. 2015 Jan 3;33(2):314-20. doi: 10.1016/j.vaccine.2014.11.026. Epub 2014 Nov 22. Vaccine. 2015. PMID: 25448110
-
Defined conjugation of glycans to the lysines of CRM197 guided by their reactivity mapping.Chembiochem. 2014 Apr 14;15(6):836-43. doi: 10.1002/cbic.201300785. Epub 2014 Mar 11. Chembiochem. 2014. PMID: 24616190
-
Designing a new antifungal glycoconjugate vaccine.Chem Soc Rev. 2013 May 21;42(10):4327-44. doi: 10.1039/c2cs35382b. Epub 2012 Dec 11. Chem Soc Rev. 2013. PMID: 23232662 Review.
-
Synthetic carbohydrate-based vaccines: challenges and opportunities.J Biomed Sci. 2020 Jan 3;27(1):9. doi: 10.1186/s12929-019-0591-0. J Biomed Sci. 2020. PMID: 31900143 Free PMC article. Review.
Cited by
-
The Hitchhiker's guide to glycoproteomics.Biochem Soc Trans. 2021 Aug 27;49(4):1643-1662. doi: 10.1042/BST20200879. Biochem Soc Trans. 2021. PMID: 34282822 Free PMC article. Review.
-
Glycovaccine Design: Optimization of Model and Antitubercular Carrier Glycosylation via Disuccinimidyl Homobifunctional Linker.Pharmaceutics. 2023 Apr 23;15(5):1321. doi: 10.3390/pharmaceutics15051321. Pharmaceutics. 2023. PMID: 37242563 Free PMC article.
-
Cytoplasmic glycoengineering of Apx toxin fragments in the development of Actinobacillus pleuropneumoniae glycoconjugate vaccines.BMC Vet Res. 2019 Jan 3;15(1):6. doi: 10.1186/s12917-018-1751-2. BMC Vet Res. 2019. PMID: 30606265 Free PMC article.
-
Synthetic Oligosaccharide-Based Vaccines Protect Mice from Clostridioides difficile Infections.ACS Chem Biol. 2019 Dec 20;14(12):2720-2728. doi: 10.1021/acschembio.9b00642. Epub 2019 Nov 19. ACS Chem Biol. 2019. PMID: 31692324 Free PMC article.
-
Alternative vaccine administration by powder injection: Needle-free dermal delivery of the glycoconjugate meningococcal group Y vaccine.PLoS One. 2017 Aug 24;12(8):e0183427. doi: 10.1371/journal.pone.0183427. eCollection 2017. PLoS One. 2017. PMID: 28837693 Free PMC article.
References
-
- Bazaka K., Crawford R. J., Nazarenko E. L. & Ivanova E. P. Bacterial Extracellular Polysaccharides. In: Bacterial Adhesion (eds Linke D., Goldman A. ). 715, 213–226. Springer: Netherlands, (2011). - PubMed
-
- Stein K. E. Thymus-independent and thymus-dependent responses to polysaccharide antigens. J of infec Dis 165 Suppl 1, S49–52 (1992). - PubMed
-
- Peltola H., Kayhty H., Sivonen A. & Makela H. Haemophilus influenzae type b capsular polysaccharide vaccine in children: a double-blind field study of 100,000 vaccinees 3 months to 5 years of age in Finland. Pediatrics 60, 730–737 (1977). - PubMed
-
- Pon R. A. Exploiting the Bacterial Surface: The Successfull Application of Glycoconjugate Vaccines In: Bacterial glycomics: current research, technology and applications (eds Reid C. W., Reid A. N., Twine S. M. ), 243–262. Horizon Scientific Press (2012).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

