Protein stability: a crystallographer's perspective
- PMID: 26841758
- PMCID: PMC4741188
- DOI: 10.1107/S2053230X15024619
Protein stability: a crystallographer's perspective
Abstract
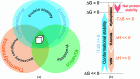
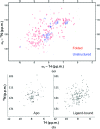
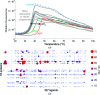
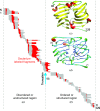
Protein stability is a topic of major interest for the biotechnology, pharmaceutical and food industries, in addition to being a daily consideration for academic researchers studying proteins. An understanding of protein stability is essential for optimizing the expression, purification, formulation, storage and structural studies of proteins. In this review, discussion will focus on factors affecting protein stability, on a somewhat practical level, particularly from the view of a protein crystallographer. The differences between protein conformational stability and protein compositional stability will be discussed, along with a brief introduction to key methods useful for analyzing protein stability. Finally, tactics for addressing protein-stability issues during protein expression, purification and crystallization will be discussed.
Keywords: crystallizability; protein crystallization; protein disorder; protein stability.
Figures

References
-
- Altmann, F., Staudacher, E., Wilson, I. B. & März, L. (1999). Insect cells as hosts for the expression of recombinant glycoproteins. Glycoconj. J. 16, 109–123. - PubMed
-
- Amarasinghe, C. & Jin, J.-P. (2015). The use of affinity tags to overcome obstacles in recombinant protein expression and purification. Protein Pept. Lett. 22, 885–892. - PubMed
-
- Anfinsen, C. B. (1973). Principles that govern the folding of protein chains. Science, 181, 223–230. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

