The tomato floral homeotic protein FBP1-like gene, SlGLO1, plays key roles in petal and stamen development
- PMID: 26842499
- PMCID: PMC4740859
- DOI: 10.1038/srep20454
The tomato floral homeotic protein FBP1-like gene, SlGLO1, plays key roles in petal and stamen development
Abstract
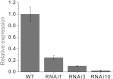
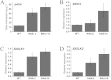
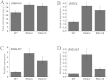
MADS-box transcription factors play important role in plant growth and development, especially floral organ identities. In our study, a MADS-box gene SlGLO1- tomato floral homeotic protein FBP1-like gene was isolated. Its tissue-specific expression profile analysis showed that SlGLO1 was highly expressed in petals and stamens. RNAi (RNA interference) repression of SlGLO1 resulted in floral organ abnormal phenotypes, including green petals with shorter size, and aberrant carpelloid stamens. SlGLO1-silenced lines are male sterile. Total chlorophyll content was increased and chlorophyll biosynthetic genes were significantly up-regulated in SlGLO1-silenced petals and stamens. Furthermore, B-class genes expression analysis indicated that the repressed function of SlGLO1 led to the enhanced expression of TAP3 and the down-regulation of TPI in the petals and stamens, while the expression of TM6 was reduced in petals and increased in stamens and carpels of SlGLO1-RNAi plants. Additionally, pollen grains of transgenic lines were aberrant and failed to germinate and tomato pollen-specific genes were down-regulated by more than 90% in SlGLO1-silenced lines. These results suggest that SlGLO1 plays important role in regulating plant floral organ and pollen development in tomato.
Figures






Similar articles
-
Silencing SlAGL6, a tomato AGAMOUS-LIKE6 lineage gene, generates fused sepal and green petal.Plant Cell Rep. 2017 Jun;36(6):959-969. doi: 10.1007/s00299-017-2129-9. Epub 2017 Mar 28. Plant Cell Rep. 2017. PMID: 28352968
-
Transcriptional and hormonal regulation of petal and stamen development by STAMENLESS, the tomato (Solanum lycopersicum L.) orthologue to the B-class APETALA3 gene.J Exp Bot. 2014 Jun;65(9):2243-56. doi: 10.1093/jxb/eru089. Epub 2014 Mar 22. J Exp Bot. 2014. PMID: 24659487 Free PMC article.
-
Functional analyses of two tomato APETALA3 genes demonstrate diversification in their roles in regulating floral development.Plant Cell. 2006 Aug;18(8):1833-45. doi: 10.1105/tpc.106.042978. Epub 2006 Jul 14. Plant Cell. 2006. PMID: 16844904 Free PMC article.
-
SlGT11 controls floral organ patterning and floral determinacy in tomato.BMC Plant Biol. 2020 Dec 14;20(1):562. doi: 10.1186/s12870-020-02760-2. BMC Plant Biol. 2020. PMID: 33317459 Free PMC article.
-
How Hormones and MADS-Box Transcription Factors Are Involved in Controlling Fruit Set and Parthenocarpy in Tomato.Genes (Basel). 2020 Nov 30;11(12):1441. doi: 10.3390/genes11121441. Genes (Basel). 2020. PMID: 33265980 Free PMC article. Review.
Cited by
-
Suppression of B function strongly supports the modified ABCE model in Tricyrtis sp. (Liliaceae).Sci Rep. 2016 Apr 15;6:24549. doi: 10.1038/srep24549. Sci Rep. 2016. PMID: 27079267 Free PMC article.
-
Silencing SlAGL6, a tomato AGAMOUS-LIKE6 lineage gene, generates fused sepal and green petal.Plant Cell Rep. 2017 Jun;36(6):959-969. doi: 10.1007/s00299-017-2129-9. Epub 2017 Mar 28. Plant Cell Rep. 2017. PMID: 28352968
-
Firming up your tomato: a natural promoter variation in a MADS-box gene is causing all-flesh tomatoes.J Exp Bot. 2022 Jan 5;73(1):1-4. doi: 10.1093/jxb/erab442. J Exp Bot. 2022. PMID: 34986230 Free PMC article.
-
Transcriptome Analysis and Screening of Genes Associated with Flower Size in Tomato (Solanum lycopersicum).Int J Mol Sci. 2022 Dec 9;23(24):15624. doi: 10.3390/ijms232415624. Int J Mol Sci. 2022. PMID: 36555271 Free PMC article.
-
Epigenetic aspects of floral homeotic genes in relation to sexual dimorphism in the dioecious plant Mercurialis annua.J Exp Bot. 2019 Nov 18;70(21):6245-6259. doi: 10.1093/jxb/erz379. J Exp Bot. 2019. PMID: 31504768 Free PMC article.
References
-
- Coen E. S. & Meyerowitz E. M. The war of the whorls: genetic interactions controlling flower development. Nature 353, 31–37 (1991). - PubMed
-
- Pelaz S., Tapia-Lopez R., Alvarez-Buylla E. R. & Yanofsky M. F. Conversion of leaves into petals in Arabidopsis. Curr. Biol. 11, 182–184 (2001). - PubMed
-
- Theissen G. et al. A short history of MADS-box genes in plants. Plant Mol. Biol. 42, 115–149 (2000). - PubMed
-
- Vrebalov J. et al. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296, 343–346 (2002). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

