Regulatory links between imprinted genes: evolutionary predictions and consequences
- PMID: 26842569
- PMCID: PMC4760173
- DOI: 10.1098/rspb.2015.2760
Regulatory links between imprinted genes: evolutionary predictions and consequences
Abstract
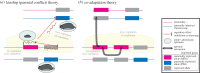
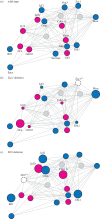
Genomic imprinting is essential for development and growth and plays diverse roles in physiology and behaviour. Imprinted genes have traditionally been studied in isolation or in clusters with respect to cis-acting modes of gene regulation, both from a mechanistic and evolutionary point of view. Recent studies in mammals, however, reveal that imprinted genes are often co-regulated and are part of a gene network involved in the control of cellular proliferation and differentiation. Moreover, a subset of imprinted genes acts in trans on the expression of other imprinted genes. Numerous studies have modulated levels of imprinted gene expression to explore phenotypic and gene regulatory consequences. Increasingly, the applied genome-wide approaches highlight how perturbation of one imprinted gene may affect other maternally or paternally expressed genes. Here, we discuss these novel findings and consider evolutionary theories that offer a rationale for such intricate interactions among imprinted genes. An evolutionary view of these trans-regulatory effects provides a novel interpretation of the logic of gene networks within species and has implications for the origin of reproductive isolation between species.
Keywords: evolution; gene network; genomic imprinting; trans regulation.
© 2016 The Authors.
Figures

References
-
- Waters AJ, Bilinski P, Eichten SR, Vaughn MW, Ross-Ibarra J, Gehring M, Springer NM. 2013. Comprehensive analysis of imprinted genes in maize reveals allelic variation for imprinting and limited conservation with other species. Proc. Natl Acad. Sci. USA 110, 19639–19644. (10.1073/pnas.1309182110) - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

