MeCP2 SUMOylation rescues Mecp2-mutant-induced behavioural deficits in a mouse model of Rett syndrome
- PMID: 26842955
- PMCID: PMC4743023
- DOI: 10.1038/ncomms10552
MeCP2 SUMOylation rescues Mecp2-mutant-induced behavioural deficits in a mouse model of Rett syndrome
Abstract
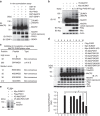
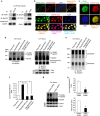
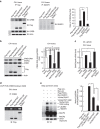
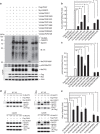
The methyl-CpG-binding protein 2 (MeCP2) gene, MECP2, is an X-linked gene encoding the MeCP2 protein, and mutations of MECP2 cause Rett syndrome (RTT). However, the molecular mechanism of MECP2-mutation-caused RTT is less known. Here we find that MeCP2 could be SUMO-modified by the E3 ligase PIAS1 at Lys-412. MeCP2 phosphorylation (at Ser-421 and Thr-308) facilitates MeCP2 SUMOylation, and MeCP2 SUMOylation is induced by NMDA, IGF-1 and CRF in the rat brain. MeCP2 SUMOylation releases CREB from the repressor complex and enhances Bdnf mRNA expression. Several MECP2 mutations identified in RTT patients show decreased MeCP2 SUMOylation. Re-expression of wild-type MeCP2 or SUMO-modified MeCP2 in Mecp2-null neurons rescues the deficits of social interaction, fear memory and LTP observed in Mecp2 conditional knockout (cKO) mice. These results together reveal an important role of MeCP2 SUMOylation in social interaction, memory and synaptic plasticity, and that abnormal MeCP2 SUMOylation is implicated in RTT.
Figures

Similar articles
-
Restoring Wnt6 signaling ameliorates behavioral deficits in MeCP2 T158A mouse model of Rett syndrome.Sci Rep. 2020 Jan 23;10(1):1074. doi: 10.1038/s41598-020-57745-w. Sci Rep. 2020. PMID: 31974426 Free PMC article.
-
Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome.J Neurosci. 2006 Jan 4;26(1):319-27. doi: 10.1523/JNEUROSCI.2623-05.2006. J Neurosci. 2006. PMID: 16399702 Free PMC article.
-
Generation and analysis of the Rett syndrome-associated MeCP2- null rat model.Yi Chuan. 2016 Nov 20;38(11):1004-1011. doi: 10.16288/j.yczz.16-180. Yi Chuan. 2016. PMID: 27867150 Review.
-
mGlu7 potentiation rescues cognitive, social, and respiratory phenotypes in a mouse model of Rett syndrome.Sci Transl Med. 2017 Aug 16;9(403):eaai7459. doi: 10.1126/scitranslmed.aai7459. Sci Transl Med. 2017. PMID: 28814546 Free PMC article.
-
Exploring the possible link between MeCP2 and oxidative stress in Rett syndrome.Free Radic Biol Med. 2015 Nov;88(Pt A):81-90. doi: 10.1016/j.freeradbiomed.2015.04.019. Epub 2015 May 8. Free Radic Biol Med. 2015. PMID: 25960047 Review.
Cited by
-
Melatonin Induction of APP Intracellular Domain 50 SUMOylation Alleviates AD through Enhanced Transcriptional Activation and Aβ Degradation.Mol Ther. 2021 Jan 6;29(1):376-395. doi: 10.1016/j.ymthe.2020.09.003. Epub 2020 Sep 5. Mol Ther. 2021. PMID: 32950104 Free PMC article.
-
Pattern decorrelation in the mouse medial prefrontal cortex enables social preference and requires MeCP2.Nat Commun. 2022 Jul 6;13(1):3899. doi: 10.1038/s41467-022-31578-9. Nat Commun. 2022. PMID: 35794118 Free PMC article.
-
Multidimensional Analysis of a Social Behavior Identifies Regression and Phenotypic Heterogeneity in a Female Mouse Model for Rett Syndrome.J Neurosci. 2024 Mar 20;44(12):e1078232023. doi: 10.1523/JNEUROSCI.1078-23.2023. J Neurosci. 2024. PMID: 38199865 Free PMC article.
-
Hsp22 pretreatment protects against LPS-induced hippocampal injury by alleviating neuroinflammation and apoptosis by regulating the NLRP3/Caspase1/IL-1β signaling pathway in mice.Aging (Albany NY). 2023 Mar 17;15(6):1977-2004. doi: 10.18632/aging.204586. Epub 2023 Mar 17. Aging (Albany NY). 2023. PMID: 36934348 Free PMC article.
-
Molecular Organization and Regulation of the Mammalian Synapse by the Post-Translational Modification SUMOylation.Cells. 2024 Feb 28;13(5):420. doi: 10.3390/cells13050420. Cells. 2024. PMID: 38474384 Free PMC article. Review.
References
-
- Amir R. E. et al. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23, 185–188 (1999). - PubMed
-
- Nan X. et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393, 386–389 (1998). - PubMed
-
- Jones P. L. et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19, 187–191 (1998). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

