Adaptive anisotropic gaussian filtering to reduce acquisition time in cardiac diffusion tensor imaging
- PMID: 26843150
- PMCID: PMC4893797
- DOI: 10.1007/s10554-016-0848-6
Adaptive anisotropic gaussian filtering to reduce acquisition time in cardiac diffusion tensor imaging
Abstract
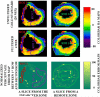
Diffusion tensor imaging (DTI) is used to quantify myocardial fiber orientation based on helical angles (HA). Accurate HA measurements require multiple excitations (NEX) and/or several diffusion encoding directions (DED). However, increasing NEX and/or DED increases acquisition time (TA). Therefore, in this study, we propose to reduce TA by implementing a 3D adaptive anisotropic Gaussian filter (AAGF) on the DTI data acquired from ex-vivo healthy and infarcted porcine hearts. DTI was performed on ex-vivo hearts [9-healthy, 3-myocardial infarction (MI)] with several combinations of DED and NEX. AAGF, mean (AVF) and median filters (MF) were applied on the primary eigenvectors of the diffusion tensor prior to HA estimation. The performance of AAGF was compared against AVF and MF. Root mean square error (RMSE), concordance correlation-coefficients and Bland-Altman's technique was used to determine optimal combination of DED and NEX that generated the best HA maps in the least possible TA. Lastly, the effect of implementing AAGF on the infarcted porcine hearts was also investigated. RMSE in HA estimation for AAGF was lower compared to AVF or MF. Post-filtering (AAGF) fewer DED and NEX were required to achieve HA maps with similar integrity as those obtained from higher NEX and/or DED. Pathological alterations caused in HA orientation in the MI model were preserved post-filtering (AAGF). Our results demonstrate that AAGF reduces TA without affecting the integrity of the myocardial microstructure.
Keywords: Anisotropic filtering; Diffusion tensor imaging (DTI); Fiber orientation; Helical angle (HA); Myocardial infarction (MI).
Figures

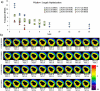
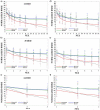
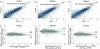


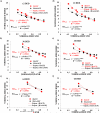
References
-
- Hunter PJ, Pullan AJ, Smaill BH. Modeling total heart function. Annu Rev Biomed Eng. 2003;5:147–77. - PubMed
-
- Kanai A, Salama G. Optical mapping reveals that repolarization spreads anisotropically and is guided by fiber orientation in guinea pig hearts. Circ Res. 1995;77(4):784–802. - PubMed
-
- Taccardi B, Lux RL, Ershler PR, et al. Anatomical architecture and electrical activity of the heart. Acta Cardiol. 1997;52(2):91–105. - PubMed
-
- LeGrice IJ, Takayama Y, Covell JW. Transverse shear along myocardial cleavage planes provides a mechanism for normal systolic wall thickening. Circ Res. 1995;77(1):182–93. - PubMed
-
- Waldman LK, Nosan D, Villarreal F, Covell JW. Relation between transmural deformation and local myofiber direction in canine left ventricle. Circ Res. 1988;63(3):550–62. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

