Analysis of B Cell Repertoire Dynamics Following Hepatitis B Vaccination in Humans, and Enrichment of Vaccine-specific Antibody Sequences
- PMID: 26844287
- PMCID: PMC4703725
- DOI: 10.1016/j.ebiom.2015.11.034
Analysis of B Cell Repertoire Dynamics Following Hepatitis B Vaccination in Humans, and Enrichment of Vaccine-specific Antibody Sequences
Abstract
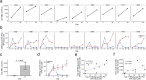
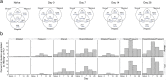
Generating a diverse B cell immunoglobulin repertoire is essential for protection against infection. The repertoire in humans can now be comprehensively measured by high-throughput sequencing. Using hepatitis B vaccination as a model, we determined how the total immunoglobulin sequence repertoire changes following antigen exposure in humans, and compared this to sequences from vaccine-specific sorted cells. Clonal sequence expansions were seen 7 days after vaccination, which correlated with vaccine-specific plasma cell numbers. These expansions caused an increase in mutation, and a decrease in diversity and complementarity-determining region 3 sequence length in the repertoire. We also saw an increase in sequence convergence between participants 14 and 21 days after vaccination, coinciding with an increase of vaccine-specific memory cells. These features allowed development of a model for in silico enrichment of vaccine-specific sequences from the total repertoire. Identifying antigen-specific sequences from total repertoire data could aid our understanding B cell driven immunity, and be used for disease diagnostics and vaccine evaluation.
Keywords: B cell repertoire; Immunoglobulin repertoire; Vaccination; mAbs.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

