Substantial Downregulation of Myogenic Transcripts in Skeletal Muscle of Atlantic Cod during the Spawning Period
- PMID: 26844771
- PMCID: PMC4742245
- DOI: 10.1371/journal.pone.0148374
Substantial Downregulation of Myogenic Transcripts in Skeletal Muscle of Atlantic Cod during the Spawning Period
Abstract
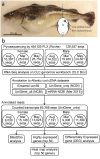
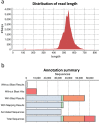
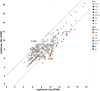
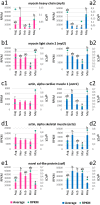
Gonadal maturation is an extremely energy consuming process for batch spawners and it is associated with a significant decrease in growth and seasonal deterioration in flesh quality. Our knowledge about the molecular mechanisms linking sexual maturation and muscle growth is still limited. In the present study, we performed RNA-Seq using 454 GS-FLX pyrosequencing in fast skeletal muscle sampled from two-year-old Atlantic cod (Gadus morhua) at representative time points throughout the reproductive cycle (August, March and May). In total, 126,937 good quality reads were obtained, with 546 nucleotide length and 52% GC content on average. RNA-Seq analysis using the CLC Genomics Workbench with the Atlantic cod reference UniGene cDNA data revealed 59,581 (46.9%) uniquely annotated reads. Pairwise comparison for expression levels identified 153 differentially expressed UniGenes between time points. Notably, we found a significant suppression of myh13 and myofibrillar gene isoforms in fast skeletal muscle during the spawning season. This study uncovered a large number of differentially expressed genes that may be influenced by gonadal maturation, thus representing a significant contribution to our limited understanding of the molecular mechanisms regulating muscle wasting and regeneration in batch spawners during their reproductive cycle.
Conflict of interest statement
Figures




Similar articles
-
Quantification of gonadotropin subunits GPalpha, FSHbeta, and LHbeta mRNA expression from Atlantic cod (Gadus morhua) throughout a reproductive cycle.Comp Biochem Physiol B Biochem Mol Biol. 2009 Jul;153(3):288-95. doi: 10.1016/j.cbpb.2009.03.011. Epub 2009 Apr 1. Comp Biochem Physiol B Biochem Mol Biol. 2009. PMID: 19344778
-
Circadian rhythmicity and photic plasticity of myosin gene transcription in fast skeletal muscle of Atlantic cod (Gadus morhua).Mar Genomics. 2014 Dec;18 Pt A:21-9. doi: 10.1016/j.margen.2014.04.011. Epub 2014 May 20. Mar Genomics. 2014. PMID: 24856374
-
Gonadal development of triploid Atlantic Cod Gadus morhua.J Fish Biol. 2011 Jun;78(7):1900-12. doi: 10.1111/j.1095-8649.2011.02955.x. Epub 2011 May 9. J Fish Biol. 2011. PMID: 21651540
-
Photoperiod effects on the expression of kisspeptin and gonadotropin genes in Atlantic cod, Gadus morhua, during first maturation.Comp Biochem Physiol A Mol Integr Physiol. 2012 Sep;163(1):82-94. doi: 10.1016/j.cbpa.2012.05.191. Epub 2012 May 18. Comp Biochem Physiol A Mol Integr Physiol. 2012. PMID: 22613785
-
Photoperiod influences growth and mll (mixed-lineage leukaemia) expression in Atlantic cod.PLoS One. 2012;7(5):e36908. doi: 10.1371/journal.pone.0036908. Epub 2012 May 9. PLoS One. 2012. PMID: 22590633 Free PMC article.
Cited by
-
Gene Expression and Phenotypic Assessment of Egg Quality across Developmental Stages of Atlantic Cod throughout the Spawning Season.Int J Mol Sci. 2024 Jul 8;25(13):7488. doi: 10.3390/ijms25137488. Int J Mol Sci. 2024. PMID: 39000593 Free PMC article.
References
-
- Bradford RG. Differential Utilization of Storage Lipids and Storage Proteins by Northwest Atlantic Herring (Clupea Harengus Harengus). J Fish Biol. 1993; 43: 811–824.
-
- Hagen O, Solberg C, Johnston IA. Activity of aspargate (cathepsin D), cysteine proteases (cathepsins B, B+L, and H), and matrix metallopeptidase (collagenase) and their influence on protein and water-holding capacity of muscle in commercially farmed Atlantic halibut (Hippoglossus hippoglossus L.). J Agric Food Chem. 2008; 56: 5953–5959. 10.1021/jf801215b - DOI - PubMed
-
- Toyohara H, Ito K, Ando M, Kinoshita M, Shimizu Y, Sakaguchi M. Effect of Maturation on Activities of Various Proteases and Protease Inhibitors in the Muscle of Ayu (Plecoglossus altivelis). Comp Biochem Physiol B. 1991; 99: 419–424. - PubMed
-
- Johnston IA, Manthri S, Alderson R, Smart A, Campbell P, Nickell D, et al. Freshwater environment affects growth rate and muscle fibre recruitment in seawater stages of Atlantic salmon (Salmo salar L.). J Exp Biol. 2003; 206: 1337–1351. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

