Effector Polymorphisms of the Sunflower Downy Mildew Pathogen Plasmopara halstedii and Their Use to Identify Pathotypes from Field Isolates
- PMID: 26845339
- PMCID: PMC4742249
- DOI: 10.1371/journal.pone.0148513
Effector Polymorphisms of the Sunflower Downy Mildew Pathogen Plasmopara halstedii and Their Use to Identify Pathotypes from Field Isolates
Abstract
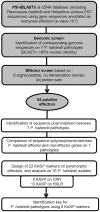
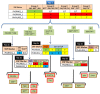
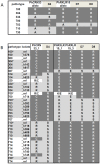
The obligate biotroph oomycete Plasmopara halstedii causes downy mildew on sunflower crop, Helianthus annuus. The breakdown of several Pl resistance genes used in sunflower hybrids over the last 25 years came along with the appearance of new Pl. halstedii isolates showing modified virulence profiles. In oomycetes, two classes of effector proteins, key players of pathogen virulence, are translocated into the host: RXLR and CRN effectors. We identified 54 putative CRN or RXLR effector genes from transcriptomic data and analyzed their genetic diversity in seven Pl. halstedii pathotypes representative of the species variability. Pl. halstedii effector genes were on average more polymorphic at both the nucleic and protein levels than random non-effector genes, suggesting a potential adaptive dynamics of pathogen virulence over the last 25 years. Twenty-two KASP (Competitive Allele Specific PCR) markers designed on polymorphic effector genes were genotyped on 35 isolates belonging to 14 Pl. halstedii pathotypes. Polymorphism analysis based on eight KASP markers aims at proposing a determination key suitable to classify the eight multi-isolate pathotypes into six groups. This is the first report of a molecular marker set able to discriminate Pl. halstedii pathotypes based on the polymorphism of pathogenicity effectors. Compared to phenotypic tests handling living spores used until now to discriminate Pl. halstedii pathotypes, this set of molecular markers constitutes a first step in faster pathotype diagnosis of Pl. halstedii isolates. Hence, emerging sunflower downy mildew isolates could be more rapidly characterized and thus, assessment of plant resistance breakdown under field conditions should be improved.
Conflict of interest statement
Figures


Similar articles
-
Sunflower resistance to multiple downy mildew pathotypes revealed by recognition of conserved effectors of the oomycete Plasmopara halstedii.Plant J. 2019 Feb;97(4):730-748. doi: 10.1111/tpj.14157. Epub 2019 Jan 7. Plant J. 2019. PMID: 30422341 Free PMC article.
-
RXLR and CRN Effectors from the Sunflower Downy Mildew Pathogen Plasmopara halstedii Induce Hypersensitive-Like Responses in Resistant Sunflower Lines.Front Plant Sci. 2016 Dec 19;7:1887. doi: 10.3389/fpls.2016.01887. eCollection 2016. Front Plant Sci. 2016. PMID: 28066456 Free PMC article.
-
The sunflower downy mildew pathogen Plasmopara halstedii.Mol Plant Pathol. 2015 Feb;16(2):109-22. doi: 10.1111/mpp.12164. Epub 2014 Dec 4. Mol Plant Pathol. 2015. PMID: 25476405 Free PMC article.
-
Sustainable and efficient control of sunflower downy mildew by means of genetic resistance: a review.Theor Appl Genet. 2022 Nov;135(11):3757-3771. doi: 10.1007/s00122-022-04038-7. Epub 2022 Jan 27. Theor Appl Genet. 2022. PMID: 35084515 Review.
-
Recent Progress in RXLR Effector Research.Mol Plant Microbe Interact. 2015 Oct;28(10):1063-72. doi: 10.1094/MPMI-01-15-0022-CR. Epub 2015 Oct 2. Mol Plant Microbe Interact. 2015. PMID: 26125490 Review.
Cited by
-
Towards low cost, multiplex clinical genotyping: 4-fluorescent Kompetitive Allele-Specific PCR and its application on pharmacogenetics.PLoS One. 2020 Mar 16;15(3):e0230445. doi: 10.1371/journal.pone.0230445. eCollection 2020. PLoS One. 2020. PMID: 32176732 Free PMC article.
-
Spatial Genetic Structure and Pathogenic Race Composition at the Field Scale in the Sunflower Downy Mildew Pathogen, Plasmopara halstedii.J Fungi (Basel). 2022 Oct 14;8(10):1084. doi: 10.3390/jof8101084. J Fungi (Basel). 2022. PMID: 36294648 Free PMC article.
-
Sunflower resistance to multiple downy mildew pathotypes revealed by recognition of conserved effectors of the oomycete Plasmopara halstedii.Plant J. 2019 Feb;97(4):730-748. doi: 10.1111/tpj.14157. Epub 2019 Jan 7. Plant J. 2019. PMID: 30422341 Free PMC article.
-
Fantastic Downy Mildew Pathogens and How to Find Them: Advances in Detection and Diagnostics.Plants (Basel). 2021 Feb 25;10(3):435. doi: 10.3390/plants10030435. Plants (Basel). 2021. PMID: 33668762 Free PMC article. Review.
-
RXLR and CRN Effectors from the Sunflower Downy Mildew Pathogen Plasmopara halstedii Induce Hypersensitive-Like Responses in Resistant Sunflower Lines.Front Plant Sci. 2016 Dec 19;7:1887. doi: 10.3389/fpls.2016.01887. eCollection 2016. Front Plant Sci. 2016. PMID: 28066456 Free PMC article.
References
-
- Nishimura M (1922) Studies in Plasmopara halstedii. J. Coll. Agric. Hokkaido Imperial University Vol XI (Pt 3): 185–210.
-
- Gulya TJ, Tourvieille de Labrouhe D, Masirevic S, Penaud A, Rashid K and Viranyi F (1998) Proposal for the standardized nomenclature and identification of races of Plasmopara halstedii (sunflower downy mildew) In: Sunflower Downy Mildew Symposium, Proceedings of Sunflower Downy Mildew Symposium, Fargo, ND, USA, pp. 130–136.
-
- Tourvieille de Labrouhe D, Walser P, Joliovot D, Roche S, Serre F, Delmotte F et al. (2012) Proposal for improvement of sunflower downy mildew race nomenclature. In: Proceedings of the 18th International Sunflower Conference, Mar del Plata, Argentina, March 2012, pp. 322–327. Paris: International Sunflower Association.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

