Reassortment of Human and Animal Rotavirus Gene Segments in Emerging DS-1-Like G1P[8] Rotavirus Strains
- PMID: 26845439
- PMCID: PMC4742054
- DOI: 10.1371/journal.pone.0148416
Reassortment of Human and Animal Rotavirus Gene Segments in Emerging DS-1-Like G1P[8] Rotavirus Strains
Abstract
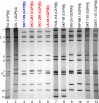
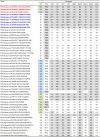
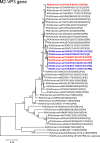
The emergence and rapid spread of novel DS-1-like G1P[8] human rotaviruses in Japan were recently reported. More recently, such intergenogroup reassortant strains were identified in Thailand, implying the ongoing spread of unusual rotavirus strains in Asia. During rotavirus surveillance in Thailand, three DS-1-like intergenogroup reassortant strains having G3P[8] (RVA/Human-wt/THA/SKT-281/2013/G3P[8] and RVA/Human-wt/THA/SKT-289/2013/G3P[8]) and G2P[8] (RVA/Human-wt/THA/LS-04/2013/G2P[8]) genotypes were identified in fecal samples from hospitalized children with acute gastroenteritis. In this study, we sequenced and characterized the complete genomes of strains SKT-281, SKT-289, and LS-04. On whole genomic analysis, all three strains exhibited unique genotype constellations including both genogroup 1 and 2 genes: G3-P[8]-I2-R2-C2-M2-A2-N2-T2-E2-H2 for strains SKT-281 and SKT-289, and G2-P[8]-I2-R2-C2-M2-A2-N2-T2-E2-H2 for strain LS-04. Except for the G genotype, the unique genotype constellation of the three strains (P[8]-I2-R2-C2-M2-A2-N2-T2-E2-H2) is commonly shared with DS-1-like G1P[8] strains. On phylogenetic analysis, nine of the 11 genes of strains SKT-281 and SKT-289 (VP4, VP6, VP1-3, NSP1-3, and NSP5) appeared to have originated from DS-1-like G1P[8] strains, while the remaining VP7 and NSP4 genes appeared to be of equine and bovine origin, respectively. Thus, strains SKT-281 and SKT-289 appeared to be reassortant strains as to DS-1-like G1P[8], animal-derived human, and/or animal rotaviruses. On the other hand, seven of the 11 genes of strain LS-04 (VP7, VP6, VP1, VP3, and NSP3-5) appeared to have originated from locally circulating DS-1-like G2P[4] human rotaviruses, while three genes (VP4, VP2, and NSP1) were assumed to be derived from DS-1-like G1P[8] strains. Notably, the remaining NSP2 gene of strain LS-04 appeared to be of bovine origin. Thus, strain LS-04 was assumed to be a multiple reassortment strain as to DS-1-like G1P[8], locally circulating DS-1-like G2P[4], bovine-like human, and/or bovine rotaviruses. Overall, the great genomic diversity among the DS-1-like G1P[8] strains seemed to have been generated through reassortment involving human and animal strains. To our knowledge, this is the first report on whole genome-based characterization of DS-1-like intergenogroup reassortant strains having G3P[8] and G2P[8] genotypes that have emerged in Thailand. Our observations will provide important insights into the evolutionary dynamics of emerging DS-1-like G1P[8] strains and related reassortant ones.
Conflict of interest statement
Figures













Similar articles
-
Full Genome Characterization of Novel DS-1-Like G8P[8] Rotavirus Strains that Have Emerged in Thailand: Reassortment of Bovine and Human Rotavirus Gene Segments in Emerging DS-1-Like Intergenogroup Reassortant Strains.PLoS One. 2016 Nov 1;11(11):e0165826. doi: 10.1371/journal.pone.0165826. eCollection 2016. PLoS One. 2016. PMID: 27802339 Free PMC article.
-
Emergence and Characterization of Unusual DS-1-Like G1P[8] Rotavirus Strains in Children with Diarrhea in Thailand.PLoS One. 2015 Nov 5;10(11):e0141739. doi: 10.1371/journal.pone.0141739. eCollection 2015. PLoS One. 2015. PMID: 26540260 Free PMC article.
-
Whole Genomic Analysis of an Unusual Human G6P[14] Rotavirus Strain Isolated from a Child with Diarrhea in Thailand: Evidence for Bovine-To-Human Interspecies Transmission and Reassortment Events.PLoS One. 2015 Sep 30;10(9):e0139381. doi: 10.1371/journal.pone.0139381. eCollection 2015. PLoS One. 2015. PMID: 26421718 Free PMC article.
-
Rotavirus Strain Trends in United States, 2009-2016: Results from the National Rotavirus Strain Surveillance System (NRSSS).Viruses. 2022 Aug 15;14(8):1775. doi: 10.3390/v14081775. Viruses. 2022. PMID: 36016397 Free PMC article. Review.
-
Rotavirus epidemiology and surveillance.Novartis Found Symp. 2001;238:125-47; discussion 147-52. doi: 10.1002/0470846534.ch9. Novartis Found Symp. 2001. PMID: 11444024 Review.
Cited by
-
Efficient packaging of HIV-1 genomes via recognition of its adenosine-rich content by a heterologous RNA-binding domain.bioRxiv [Preprint]. 2025 May 1:2025.05.01.651415. doi: 10.1101/2025.05.01.651415. bioRxiv. 2025. PMID: 40654762 Free PMC article. Preprint.
-
Molecular characterization of human group A rotavirus genotypes circulating in Rawalpindi, Islamabad, Pakistan during 2015-2016.PLoS One. 2019 Jul 30;14(7):e0220387. doi: 10.1371/journal.pone.0220387. eCollection 2019. PLoS One. 2019. PMID: 31361761 Free PMC article.
-
Molecular characterisation of rotavirus strains detected during a clinical trial of the human neonatal rotavirus vaccine (RV3-BB) in Indonesia.Vaccine. 2018 Sep 18;36(39):5872-5878. doi: 10.1016/j.vaccine.2018.08.027. Epub 2018 Aug 23. Vaccine. 2018. PMID: 30145099 Free PMC article. Clinical Trial.
-
Genetic characterization of rotavirus A strains circulating in children under 5 years of age with acute gastroenteritis in Tehran, Iran, in 2023-2024: dissemination of the emerging equine-like G3P[8]-I2-E2 DS-1-like strains.J Gen Virol. 2025 Mar;106(3):002088. doi: 10.1099/jgv.0.002088. J Gen Virol. 2025. PMID: 40100090 Free PMC article.
-
First Detection of DS-1-like G1P[8] Double-gene Reassortant Rotavirus Strains on The American Continent, Brazil, 2013.Sci Rep. 2019 Feb 18;9(1):2210. doi: 10.1038/s41598-019-38703-7. Sci Rep. 2019. PMID: 30778110 Free PMC article.
References
-
- Tate JE, Burton AH, Boschi-Pinto C, Steele AD, Duque J, Parashar UD, et al. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: a systematic review and meta-analysis. Lancet Infect Dis. 2012;12: 136–141. 10.1016/S1473-3099(11)70253-5 - DOI - PubMed
-
- Kahn G, Fitzwater S, Tate J, Kang G, Ganguly N, Nair G, et al. Epidemiology and prospects for prevention of rotavirus disease in India. Indian Pediatr. 2012;49: 467–474. - PubMed
-
- Estes MK, Greenberg HB. Rotaviruses In: Knipe DM, Howley PM, editors. Fields Virology. 6th ed. Philadelphia: Lippincott Williams & Wilkins; 2013. pp. 1347–1401.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

