Genomic Modifiers of Natural Killer Cells, Immune Responsiveness and Lymphoid Tissue Remodeling Together Increase Host Resistance to Viral Infection
- PMID: 26845690
- PMCID: PMC4742223
- DOI: 10.1371/journal.ppat.1005419
Genomic Modifiers of Natural Killer Cells, Immune Responsiveness and Lymphoid Tissue Remodeling Together Increase Host Resistance to Viral Infection
Abstract
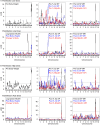
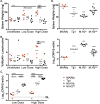
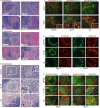
The MHC class I D(k) molecule supplies vital host resistance during murine cytomegalovirus (MCMV) infection. Natural killer (NK) cells expressing the Ly49G2 inhibitory receptor, which specifically binds D(k), are required to control viral spread. The extent of D(k)-dependent host resistance, however, differs significantly amongst related strains of mice, C57L and MA/My. As a result, we predicted that relatively small-effect modifier genetic loci might together shape immune cell features, NK cell reactivity, and the host immune response to MCMV. A robust D(k)-dependent genetic effect, however, has so far hindered attempts to identify additional host resistance factors. Thus, we applied genomic mapping strategies and multicolor flow cytometric analysis of immune cells in naive and virus-infected hosts to identify genetic modifiers of the host immune response to MCMV. We discovered and validated many quantitative trait loci (QTL); these were mapped to at least 19 positions on 16 chromosomes. Intriguingly, one newly discovered non-MHC locus (Cmv5) controlled splenic NK cell accrual, secondary lymphoid organ structure, and lymphoid follicle development during MCMV infection. We infer that Cmv5 aids host resistance to MCMV infection by expanding NK cells needed to preserve and protect essential tissue structural elements, to enhance lymphoid remodeling and to increase viral clearance in spleen.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Multiparametric analysis of host response to murine cytomegalovirus in MHC class I-disparate mice reveals primacy of Dk-licensed Ly49G2+ NK cells in viral control.J Immunol. 2013 Nov 1;191(9):4709-19. doi: 10.4049/jimmunol.1301388. Epub 2013 Sep 25. J Immunol. 2013. PMID: 24068668 Free PMC article.
-
NK gene complex and chromosome 19 loci enhance MHC resistance to murine cytomegalovirus infection.Immunogenetics. 2009 Dec;61(11-12):755-64. doi: 10.1007/s00251-009-0400-0. Epub 2010 Jan 5. Immunogenetics. 2009. PMID: 19820922 Free PMC article.
-
Genome-Wide Exome Analysis of Cmv5-Disparate Mouse Strains that Differ in Host Resistance to Murine Cytomegalovirus Infection.G3 (Bethesda). 2017 Jun 7;7(6):1979-1984. doi: 10.1534/g3.117.042531. G3 (Bethesda). 2017. PMID: 28450376 Free PMC article.
-
NK cell recognition of mouse cytomegalovirus-infected cells.Curr Top Microbiol Immunol. 2006;298:183-206. doi: 10.1007/3-540-27743-9_10. Curr Top Microbiol Immunol. 2006. PMID: 16329187 Review.
-
Natural killer cell licensing during viral infection.Adv Exp Med Biol. 2011;780:37-44. doi: 10.1007/978-1-4419-5632-3_4. Adv Exp Med Biol. 2011. PMID: 21842363 Review.
Cited by
-
Licensing Natural Killers for Antiviral Immunity.Pathogens. 2021 Jul 19;10(7):908. doi: 10.3390/pathogens10070908. Pathogens. 2021. PMID: 34358058 Free PMC article. Review.
-
Dose of Retroviral Infection Determines Induction of Antiviral NK Cell Responses.J Virol. 2017 Oct 27;91(22):e01122-17. doi: 10.1128/JVI.01122-17. Print 2017 Nov 15. J Virol. 2017. PMID: 28904191 Free PMC article.
-
Atraumatic Splenic Rupture Unveiling Mumps With an Underlying B-cell Lymphoid Hyperplasia: A Diagnostic Conundrum.Cureus. 2024 Oct 29;16(10):e72671. doi: 10.7759/cureus.72671. eCollection 2024 Oct. Cureus. 2024. PMID: 39478769 Free PMC article.
-
Multiple Immune and Genetic Mechanisms Contribute to Cmv5s-Driven Susceptibility and Tissue Damage during Acute Murine Cytomegalovirus Infection.J Immunol. 2024 Mar 1;212(5):813-824. doi: 10.4049/jimmunol.2300648. J Immunol. 2024. PMID: 38224204 Free PMC article.
-
Natural selection for killer receptors and their MHC class I ligands: In pursuit of gene pairs that fit well in tandem.J Leukoc Biol. 2019 Mar;105(3):489-495. doi: 10.1002/JLB.2RI0818-315R. Epub 2018 Nov 30. J Leukoc Biol. 2019. PMID: 30500089 Free PMC article. Review.
References
-
- Theiler M (1930) Susceptibility of White Mice to the Virus of Yellow Fever. Science 71: 367. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

