The Symbiotic Performance of Chickpea Rhizobia Can Be Improved by Additional Copies of the clpB Chaperone Gene
- PMID: 26845770
- PMCID: PMC4741418
- DOI: 10.1371/journal.pone.0148221
The Symbiotic Performance of Chickpea Rhizobia Can Be Improved by Additional Copies of the clpB Chaperone Gene
Abstract
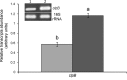
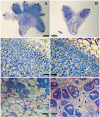
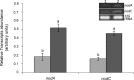
The ClpB chaperone is known to be involved in bacterial stress response. Moreover, recent studies suggest that this protein has also a role in the chickpea-rhizobia symbiosis. In order to improve both stress tolerance and symbiotic performance of a chickpea microsymbiont, the Mesorhizobium mediterraneum UPM-Ca36T strain was genetically transformed with pPHU231 containing an extra-copy of the clpB gene. To investigate if the clpB-transformed strain displays an improved stress tolerance, bacterial growth was evaluated under heat and acid stress conditions. In addition, the effect of the extra-copies of the clpB gene in the symbiotic performance was evaluated using plant growth assays (hydroponic and pot trials). The clpB-transformed strain is more tolerant to heat shock than the strain transformed with pPHU231, supporting the involvement of ClpB in rhizobia heat shock tolerance. Both plant growth assays showed that ClpB has an important role in chickpea-rhizobia symbiosis. The nodulation kinetics analysis showed a higher rate of nodule appearance with the clpB-transformed strain. This strain also induced a greater number of nodules and, more notably, its symbiotic effectiveness increased ~60% at pH5 and 83% at pH7, compared to the wild-type strain. Furthermore, a higher frequency of root hair curling was also observed in plants inoculated with the clpB-transformed strain, compared to the wild-type strain. The superior root hair curling induction, nodulation ability and symbiotic effectiveness of the clpB-transformed strain may be explained by an increased expression of symbiosis genes. Indeed, higher transcript levels of the nodulation genes nodA and nodC (~3 folds) were detected in the clpB-transformed strain. The improvement of rhizobia by addition of extra-copies of the clpB gene may be a promising strategy to obtain strains with enhanced stress tolerance and symbiotic effectiveness, thus contributing to their success as crop inoculants, particularly under environmental stresses. This is the first report on the successful improvement of a rhizobium with a chaperone gene.
Conflict of interest statement
Figures




Similar articles
-
Can stress response genes be used to improve the symbiotic performance of rhizobia?AIMS Microbiol. 2017 May 26;3(3):365-382. doi: 10.3934/microbiol.2017.3.365. eCollection 2017. AIMS Microbiol. 2017. PMID: 31294167 Free PMC article. Review.
-
A ClpB chaperone knockout mutant of Mesorhizobium ciceri shows a delay in the root nodulation of chickpea plants.Mol Plant Microbe Interact. 2012 Dec;25(12):1594-604. doi: 10.1094/MPMI-05-12-0140-R. Mol Plant Microbe Interact. 2012. PMID: 23134119
-
Genotypic alteration and competitive nodulation of Mesorhizobium muleiense against exotic chickpea rhizobia in alkaline soils.Syst Appl Microbiol. 2014 Oct;37(7):520-4. doi: 10.1016/j.syapm.2014.07.004. Epub 2014 Jul 21. Syst Appl Microbiol. 2014. PMID: 25123757
-
Transcriptional analysis of major chaperone genes in salt-tolerant and salt-sensitive mesorhizobia.Microbiol Res. 2012 Dec 20;167(10):623-9. doi: 10.1016/j.micres.2012.01.006. Epub 2012 Feb 23. Microbiol Res. 2012. PMID: 22364959
-
Nodulation outer proteins: double-edged swords of symbiotic rhizobia.Biochem J. 2015 Sep 15;470(3):263-74. doi: 10.1042/BJ20150518. Biochem J. 2015. PMID: 26341483 Review.
Cited by
-
Rhizosphere microbiome influence on tomato growth under low-nutrient settings.FEMS Microbiol Ecol. 2025 Feb 20;101(3):fiaf019. doi: 10.1093/femsec/fiaf019. FEMS Microbiol Ecol. 2025. PMID: 39999861 Free PMC article.
-
Can stress response genes be used to improve the symbiotic performance of rhizobia?AIMS Microbiol. 2017 May 26;3(3):365-382. doi: 10.3934/microbiol.2017.3.365. eCollection 2017. AIMS Microbiol. 2017. PMID: 31294167 Free PMC article. Review.
-
traG Gene Is Conserved across Mesorhizobium spp. Able to Nodulate the Same Host Plant and Expressed in Response to Root Exudates.Biomed Res Int. 2019 Jan 30;2019:3715271. doi: 10.1155/2019/3715271. eCollection 2019. Biomed Res Int. 2019. PMID: 30834262 Free PMC article.
-
Harnessing chickpea bacterial endophytes for improved plant health and fitness.AIMS Microbiol. 2024 Jul 8;10(3):489-506. doi: 10.3934/microbiol.2024024. eCollection 2024. AIMS Microbiol. 2024. PMID: 39219751 Free PMC article. Review.
-
An overview of heat stress in Chickpea (Cicer arietinum L.): effects, mechanisms and diverse molecular breeding approaches for enhancing resilience and productivity.Mol Breed. 2025 Jan 21;45(2):18. doi: 10.1007/s11032-025-01538-4. eCollection 2025 Feb. Mol Breed. 2025. PMID: 39850651 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

