Histone H4 acetylation and the epigenetic reader Brd4 are critical regulators of pluripotency in embryonic stem cells
- PMID: 26847871
- PMCID: PMC4740988
- DOI: 10.1186/s12864-016-2414-y
Histone H4 acetylation and the epigenetic reader Brd4 are critical regulators of pluripotency in embryonic stem cells
Abstract
Background: Pluripotent cells can be differentiated into many different cell types in vitro. Successful differentiation is guided in large part by epigenetic reprogramming and regulation of critical gene expression patterns. Recent genome-wide studies have identified the distribution of different histone-post-translational modifications (PTMs) in various conditions and during cellular differentiation. However, our understanding of the abundance of histone PTMs and their regulatory mechanisms still remain unknown.
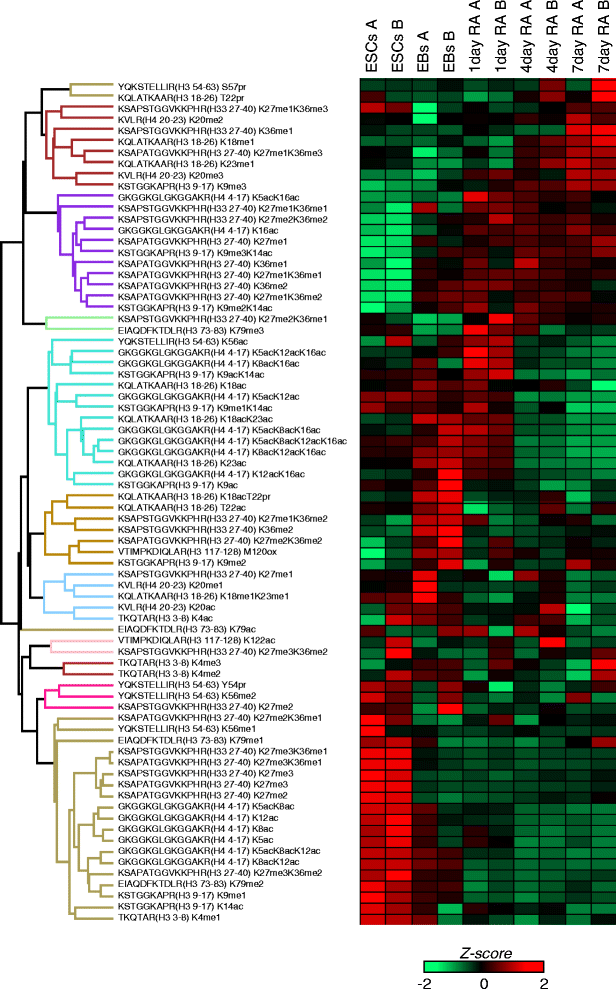
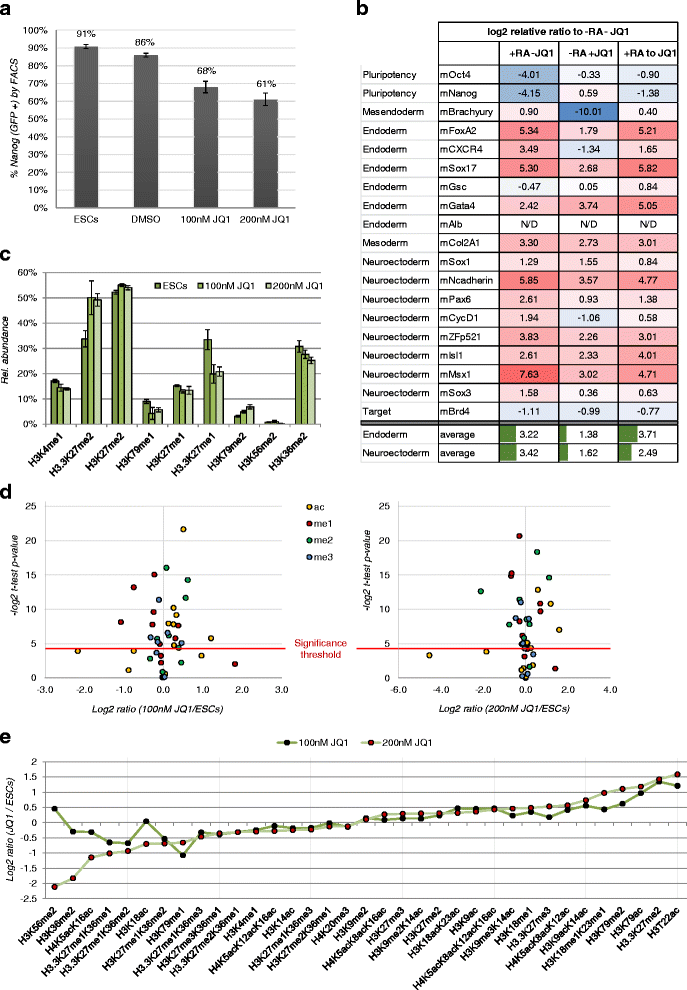
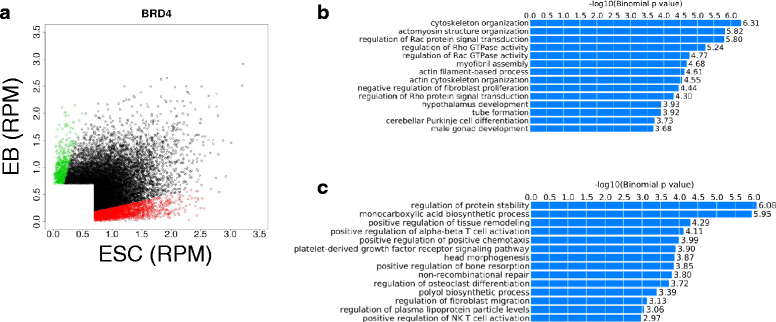
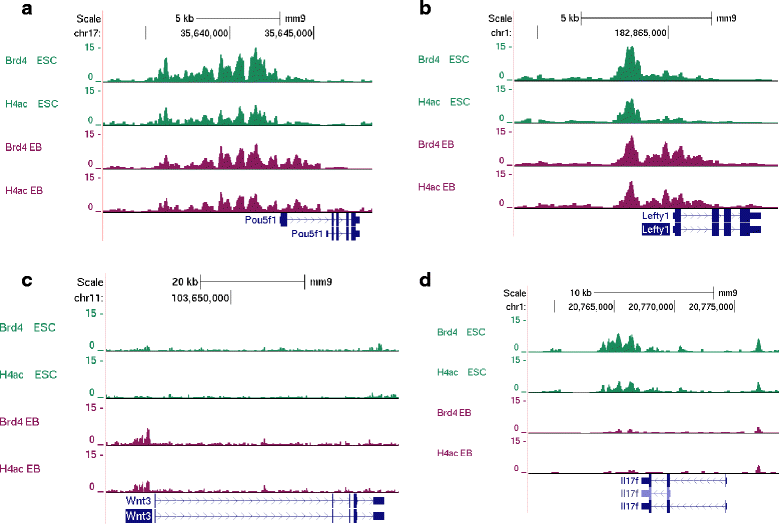
Results: Here, we present a quantitative and comprehensive study of the abundance levels of histone PTMs during the differentiation of mouse embryonic stem cells (ESCs) using mass spectrometry (MS). We observed dynamic changes of histone PTMs including increased H3K9 methylation levels in agreement with previously reported results. More importantly, we found a global decrease of multiply acetylated histone H4 peptides. Brd4 targets acetylated H4 with a strong affinity to multiply modified H4 acetylation sites. We observed that the protein levels of Brd4 decreased upon differentiation together with global histone H4 acetylation. Inhibition of Brd4:histone H4 interaction by the BET domain inhibitor (+)-JQ1 in ESCs results in enhanced differentiation to the endodermal lineage, by disrupting the protein abundance dynamics. Genome-wide ChIP-seq mapping showed that Brd4 and H4 acetylation are co-occupied in the genome, upstream of core pluripotency genes such as Oct4 and Nanog in ESCs and lineage-specific genes in embryoid bodies (EBs).
Conclusions: Together, our data demonstrate the fundamental role of Brd4 in monitoring cell differentiation through its interaction with acetylated histone marks and disruption of Brd4 may cause aberrant differentiation.
Figures


Similar articles
-
Nanog requires BRD4 to maintain murine embryonic stem cell pluripotency and is suppressed by bromodomain inhibitor JQ1 together with Lefty1.Stem Cells Dev. 2015 Apr 1;24(7):879-91. doi: 10.1089/scd.2014.0302. Epub 2014 Dec 17. Stem Cells Dev. 2015. PMID: 25393219 Free PMC article.
-
Brd4's Bromodomains Mediate Histone H3 Acetylation and Chromatin Remodeling in Pluripotent Cells through P300 and Brg1.Cell Rep. 2018 Nov 13;25(7):1756-1771. doi: 10.1016/j.celrep.2018.10.003. Cell Rep. 2018. PMID: 30428346
-
Cell differentiation along multiple pathways accompanied by changes in histone acetylation status.Biochem Cell Biol. 2014 Apr;92(2):85-93. doi: 10.1139/bcb-2013-0082. Epub 2014 Jan 17. Biochem Cell Biol. 2014. PMID: 24697692
-
Histone variants as emerging regulators of embryonic stem cell identity.Epigenetics. 2015;10(7):563-73. doi: 10.1080/15592294.2015.1053682. Epigenetics. 2015. PMID: 26114724 Free PMC article. Review.
-
Returning to the stem state: epigenetics of recapitulating pre-differentiation chromatin structure.Bioessays. 2010 Sep;32(9):791-9. doi: 10.1002/bies.201000033. Bioessays. 2010. PMID: 20652894 Review.
Cited by
-
Resolving Geroplasticity to the Balance of Rejuvenins and Geriatrins.Aging Dis. 2022 Dec 1;13(6):1664-1714. doi: 10.14336/AD.2022.0414. eCollection 2022 Dec 1. Aging Dis. 2022. PMID: 36465174 Free PMC article.
-
A Novel Epi-drug Therapy Based on the Suppression of BET Family Epigenetic Readers.Yale J Biol Med. 2017 Mar 29;90(1):63-71. eCollection 2017 Mar. Yale J Biol Med. 2017. PMID: 28356894 Free PMC article. Review.
-
A crucial role for dynamic expression of components encoding the negative arm of the circadian clock.Nat Commun. 2023 Jun 8;14(1):3371. doi: 10.1038/s41467-023-38817-7. Nat Commun. 2023. PMID: 37291101 Free PMC article.
-
Proteome-wide acetylation dynamics in human cells.Sci Rep. 2017 Aug 31;7(1):10296. doi: 10.1038/s41598-017-09918-3. Sci Rep. 2017. PMID: 28860605 Free PMC article.
-
Facultative dosage compensation of developmental genes on autosomes in Drosophila and mouse embryonic stem cells.Nat Commun. 2018 Sep 7;9(1):3626. doi: 10.1038/s41467-018-05642-2. Nat Commun. 2018. PMID: 30194291 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

