Maternal transcription of non-protein coding RNAs from the PWS-critical region rescues growth retardation in mice
- PMID: 26848093
- PMCID: PMC4742849
- DOI: 10.1038/srep20398
Maternal transcription of non-protein coding RNAs from the PWS-critical region rescues growth retardation in mice
Abstract
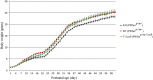
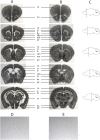
Prader-Willi syndrome (PWS) is a neurogenetic disorder caused by loss of paternally expressed genes on chromosome 15q11-q13. The PWS-critical region (PWScr) contains an array of non-protein coding IPW-A exons hosting intronic SNORD116 snoRNA genes. Deletion of PWScr is associated with PWS in humans and growth retardation in mice exhibiting ~15% postnatal lethality in C57BL/6 background. Here we analysed a knock-in mouse containing a 5'HPRT-LoxP-Neo(R) cassette (5'LoxP) inserted upstream of the PWScr. When the insertion was inherited maternally in a paternal PWScr-deletion mouse model (PWScr(p-/m5'LoxP)), we observed compensation of growth retardation and postnatal lethality. Genomic methylation pattern and expression of protein-coding genes remained unaltered at the PWS-locus of PWScr(p-/m5'LoxP) mice. Interestingly, ubiquitous Snord116 and IPW-A exon transcription from the originally silent maternal chromosome was detected. In situ hybridization indicated that PWScr(p-/m5'LoxP) mice expressed Snord116 in brain areas similar to wild type animals. Our results suggest that the lack of PWScr RNA expression in certain brain areas could be a primary cause of the growth retardation phenotype in mice. We propose that activation of disease-associated genes on imprinted regions could lead to general therapeutic strategies in associated diseases.
Figures



References
-
- Runte M. et al. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet 10, 2687–2700 (2001). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

