Detection rate of actionable mutations in diverse cancers using a biopsy-free (blood) circulating tumor cell DNA assay
- PMID: 26848768
- PMCID: PMC4891078
- DOI: 10.18632/oncotarget.7110
Detection rate of actionable mutations in diverse cancers using a biopsy-free (blood) circulating tumor cell DNA assay
Abstract
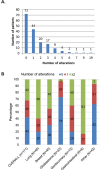
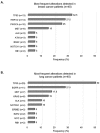
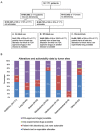
Analysis of cell-free DNA using next-generation sequencing (NGS) is a powerful tool for the detection/monitoring of alterations present in circulating tumor DNA (ctDNA). Plasma extracted from 171 patients with a variety of cancers was analyzed for ctDNA (54 genes and copy number variants (CNVs) in three genes (EGFR, ERBB2 and MET)). The most represented cancers were lung (23%), breast (23%), and glioblastoma (19%). Ninety-nine patients (58%) had at least one detectable alteration. The most frequent alterations were TP53 (29.8%), followed by EGFR (17.5%), MET (10.5%), PIK3CA (7%), and NOTCH1 (5.8%). In contrast, of 222 healthy volunteers, only one had an aberration (TP53). Ninety patients with non-brain tumors had a discernible aberration (65% of 138 patients; in 70% of non-brain tumor patients with an alteration, the anomaly was potentially actionable). Interestingly, nine of 33 patients (27%) with glioblastoma had an alteration (6/33 (18%) potentially actionable). Overall, sixty-nine patients had potentially actionable alterations (40% of total; 69.7% of patients (69/99) with alterations); 68 patients (40% of total; 69% of patients with alterations), by a Food and Drug Administration (FDA) approved drug. In summary, 65% of diverse cancers (as well as 27% of glioblastomas) had detectable ctDNA aberration(s), with the majority theoretically actionable by an approved agent.
Keywords: actionable alteration; cancer; ctDNA; liquid biopsy; personalized therapy.
Conflict of interest statement
Dr. Kurzrock has research funding from Genentech, Merck Serono, Pfizer, and Foundation Medicine, as well as consultant fees from Sequenom and is the founder of RScueRX, Inc. Ms. Banks and Dr. Lanman are employees of Guardant Health, Inc. Dr. Talasaz is co-founder and employee of Guardant Health, Inc. The other authors have nothing to disclose.
Figures

References
-
- Tsimberidou A-M, Iskander NG, Hong DS, Wheler JJ, Falchook GS, Fu S, Piha-Paul S, Naing A, Janku F, Luthra R, Ye Y, Wen S, Berry D, Kurzrock R. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res. 2012 Nov 15;18:6373–83. - PMC - PubMed
-
- Schwaederle M, Parker BA, Schwab RB, Fanta PT, Boles SG, Daniels GA, Bazhenova LA, Subramanian R, Coutinho AC, Ojeda-Fournier H, Datnow B, Webster NJ, Lippman SM, Kurzrock R. Molecular tumor board: the University of California-San Diego Moores Cancer Center experience. The Oncologist. 2014 Jun;19:631–6.. - PMC - PubMed
-
- Von Hoff DD, Stephenson JJ, Jr., Rosen P, Loesch DM, Borad MJ, Anthony S, Jameson G, Brown S, Cantafio N, Richards DA, Fitch TR, Wasserman E, Fernandez C, et al. Pilot study using molecular profiling of patients' tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol. 2010 Nov 20;28:4877–83. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

