Recruitment of Saccharomyces cerevisiae Cmr1/Ydl156w to Coding Regions Promotes Transcription Genome Wide
- PMID: 26848854
- PMCID: PMC4744024
- DOI: 10.1371/journal.pone.0148897
Recruitment of Saccharomyces cerevisiae Cmr1/Ydl156w to Coding Regions Promotes Transcription Genome Wide
Abstract
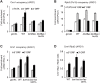
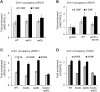
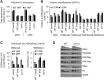
Cmr1 (changed mutation rate 1) is a largely uncharacterized nuclear protein that has recently emerged in several global genetic interaction and protein localization studies. It clusters with proteins involved in DNA damage and replication stress response, suggesting a role in maintaining genome integrity. Under conditions of proteasome inhibition or replication stress, this protein localizes to distinct sub-nuclear foci termed as intranuclear quality control (INQ) compartments, which sequester proteins for their subsequent degradation. Interestingly, it also interacts with histones, chromatin remodelers and modifiers, as well as with proteins involved in transcription including subunits of RNA Pol I and Pol III, but not with those of Pol II. It is not known whether Cmr1 plays a role in regulating transcription of Pol II target genes. Here, we show that Cmr1 is recruited to the coding regions of transcribed genes of S. cerevisiae. Cmr1 occupancy correlates with the Pol II occupancy genome-wide, indicating that it is recruited to coding sequences in a transcription-dependent manner. Cmr1-enriched genes include Gcn4 targets and ribosomal protein genes. Furthermore, our results show that Cmr1 recruitment to coding sequences is stimulated by Pol II CTD kinase, Kin28, and the histone deacetylases, Rpd3 and Hos2. Finally, our genome-wide analyses implicate Cmr1 in regulating Pol II occupancy at transcribed coding sequences. However, it is dispensable for maintaining co-transcriptional histone occupancy and histone modification (acetylation and methylation). Collectively, our results show that Cmr1 facilitates transcription by directly engaging with transcribed coding regions.
Conflict of interest statement
Figures



Similar articles
-
Acetylation-Dependent Recruitment of the FACT Complex and Its Role in Regulating Pol II Occupancy Genome-Wide in Saccharomyces cerevisiae.Genetics. 2018 Jul;209(3):743-756. doi: 10.1534/genetics.118.300943. Epub 2018 Apr 25. Genetics. 2018. PMID: 29695490 Free PMC article.
-
The RSC complex localizes to coding sequences to regulate Pol II and histone occupancy.Mol Cell. 2014 Dec 4;56(5):653-66. doi: 10.1016/j.molcel.2014.10.002. Epub 2014 Nov 6. Mol Cell. 2014. PMID: 25457164 Free PMC article.
-
Histone deacetylases and phosphorylated polymerase II C-terminal domain recruit Spt6 for cotranscriptional histone reassembly.Mol Cell Biol. 2014 Nov 15;34(22):4115-29. doi: 10.1128/MCB.00695-14. Epub 2014 Sep 2. Mol Cell Biol. 2014. PMID: 25182531 Free PMC article.
-
Genome-wide RNA polymerase II: not genes only!Trends Biochem Sci. 2008 Jun;33(6):265-73. doi: 10.1016/j.tibs.2008.04.006. Epub 2008 May 6. Trends Biochem Sci. 2008. PMID: 18467100 Review.
-
A site to remember: H3K36 methylation a mark for histone deacetylation.Mutat Res. 2007 May 1;618(1-2):130-4. doi: 10.1016/j.mrfmmm.2006.08.014. Epub 2007 Jan 21. Mutat Res. 2007. PMID: 17346757 Review.
Cited by
-
The role of WDR76 protein in human diseases.Bosn J Basic Med Sci. 2021 Oct 1;21(5):528-534. doi: 10.17305/bjbms.2020.5506. Bosn J Basic Med Sci. 2021. PMID: 33714259 Free PMC article. Review.
-
Selective aggregation of the splicing factor Hsh155 suppresses splicing upon genotoxic stress.J Cell Biol. 2017 Dec 4;216(12):4027-4040. doi: 10.1083/jcb.201612018. Epub 2017 Oct 4. J Cell Biol. 2017. PMID: 28978642 Free PMC article.
-
HDA-2-Containing Complex Is Required for Activation of Catalase-3 Expression in Neurospora crassa.mBio. 2022 Aug 30;13(4):e0135122. doi: 10.1128/mbio.01351-22. Epub 2022 Jun 14. mBio. 2022. PMID: 35699373 Free PMC article.
-
Biochemical Reduction of the Topology of the Diverse WDR76 Protein Interactome.J Proteome Res. 2019 Sep 6;18(9):3479-3491. doi: 10.1021/acs.jproteome.9b00373. Epub 2019 Aug 9. J Proteome Res. 2019. PMID: 31353912 Free PMC article.
-
WDR76 Co-Localizes with Heterochromatin Related Proteins and Rapidly Responds to DNA Damage.PLoS One. 2016 Jun 1;11(6):e0155492. doi: 10.1371/journal.pone.0155492. eCollection 2016. PLoS One. 2016. PMID: 27248496 Free PMC article.
References
-
- Patterson AV, Saunders MP, Chinje EC, Patterson LH, Stratford IJ. Enzymology of tirapazamine metabolism: a review. Anticancer Drug Des. 1998;13(6):541–73. Epub 1998/10/02. . - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

