Synonymous Codons Direct Cotranslational Folding toward Different Protein Conformations
- PMID: 26849192
- PMCID: PMC4745992
- DOI: 10.1016/j.molcel.2016.01.008
Synonymous Codons Direct Cotranslational Folding toward Different Protein Conformations
Abstract
In all genomes, most amino acids are encoded by more than one codon. Synonymous codons can modulate protein production and folding, but the mechanism connecting codon usage to protein homeostasis is not known. Here we show that synonymous codon variants in the gene encoding gamma-B crystallin, a mammalian eye-lens protein, modulate the rates of translation and cotranslational folding of protein domains monitored in real time by Förster resonance energy transfer and fluorescence-intensity changes. Gamma-B crystallins produced from mRNAs with changed codon bias have the same amino acid sequence but attain different conformations, as indicated by altered in vivo stability and in vitro protease resistance. 2D NMR spectroscopic data suggest that structural differences are associated with different cysteine oxidation states of the purified proteins, providing a link between translation, folding, and the structures of isolated proteins. Thus, synonymous codons provide a secondary code for protein folding in the cell.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
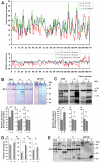



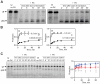

References
-
- Bloemendal H, de Jong W, Jaenicke R, Lubsen NH, Slingsby C, Tardieu A. Ageing and vision: structure, stability and function of lens crystallins. Prog. Biophys. Mol. Biol. 2004;86:407–485. - PubMed
-
- Buchan JR, Stansfield I. Halting a cellular production line: responses to ribosomal pausing during translation. Biol. Cell. 2007;99:475–487. - PubMed
-
- Cabrita LD, Dobson CM, Christodoulou J. Protein folding on the ribosome. Curr. Opin. Struct. Biol. 2010;20:33–45. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
