Identifying Patient-Specific Epstein-Barr Nuclear Antigen-1 Genetic Variation and Potential Autoreactive Targets Relevant to Multiple Sclerosis Pathogenesis
- PMID: 26849221
- PMCID: PMC4744032
- DOI: 10.1371/journal.pone.0147567
Identifying Patient-Specific Epstein-Barr Nuclear Antigen-1 Genetic Variation and Potential Autoreactive Targets Relevant to Multiple Sclerosis Pathogenesis
Abstract
Background: Epstein-Barr virus (EBV) infection represents a major environmental risk factor for multiple sclerosis (MS), with evidence of selective expansion of Epstein-Barr Nuclear Antigen-1 (EBNA1)-specific CD4+ T cells that cross-recognize MS-associated myelin antigens in MS patients. HLA-DRB1*15-restricted antigen presentation also appears to determine susceptibility given its role as a dominant risk allele. In this study, we have utilised standard and next-generation sequencing techniques to investigate EBNA-1 sequence variation and its relationship to HLA-DR15 binding affinity, as well as examining potential cross-reactive immune targets within the central nervous system proteome.
Methods: Sanger sequencing was performed on DNA isolated from peripheral blood samples from 73 Western Australian MS cases, without requirement for primary culture, with additional FLX 454 Roche sequencing in 23 samples to identify low-frequency variants. Patient-derived viral sequences were used to predict HLA-DRB1*1501 epitopes (NetMHCII, NetMHCIIpan) and candidates were evaluated for cross recognition with human brain proteins.
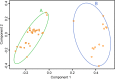
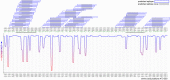
Results: EBNA-1 sequence variation was limited, with no evidence of multiple viral strains and only low levels of variation identified by FLX technology (8.3% nucleotide positions at a 1% cut-off). In silico epitope mapping revealed two known HLA-DRB1*1501-restricted epitopes ('AEG': aa 481-496 and 'MVF': aa 562-577), and two putative epitopes between positions 502-543. We identified potential cross-reactive targets involving a number of major myelin antigens including experimentally confirmed HLA-DRB1*15-restricted epitopes as well as novel candidate antigens within myelin and paranodal assembly proteins that may be relevant to MS pathogenesis.
Conclusions: This study demonstrates the feasibility of obtaining autologous EBNA-1 sequences directly from buffy coat samples, and confirms divergence of these sequences from standard laboratory strains. This approach has identified a number of immunogenic regions of EBNA-1 as well as known and novel targets for autoreactive HLA-DRB1*15-restricted T cells within the central nervous system that could arise as a result of cross-reactivity with EBNA-1-specific immune responses.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

