Contributions of ancestral inter-species recombination to the genetic diversity of extant Streptomyces lineages
- PMID: 26849310
- PMCID: PMC4918437
- DOI: 10.1038/ismej.2015.230
Contributions of ancestral inter-species recombination to the genetic diversity of extant Streptomyces lineages
Abstract
Streptomyces species produce many important antibiotics and have a crucial role in soil nutrient cycling. However, their evolutionary history remains poorly characterized. We have evaluated the impact of homologous recombination on the evolution of Streptomyces using multi-locus sequence analysis of 234 strains that represent at least 11 species clusters. Evidence of inter-species recombination is widespread but not uniform within the genus and levels of mosaicism vary between species clusters. Most phylogenetically incongruent loci are monophyletic at the scale of species clusters and their subclades, suggesting that these recombination events occurred in shared ancestral lineages. Further investigation of two mosaic species clusters suggests that genes acquired by inter-species recombination may have become fixed in these lineages during periods of demographic expansion; implicating a role for phylogeography in determining contemporary patterns of genetic diversity. Only by examining the phylogeny at the scale of the genus is apparent that widespread phylogenetically incongruent loci in Streptomyces are derived from a far smaller number of ancestral inter-species recombination events.
Figures
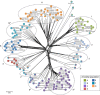
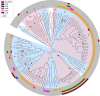


Similar articles
-
A Latitudinal Diversity Gradient in Terrestrial Bacteria of the Genus Streptomyces.mBio. 2016 Apr 12;7(2):e02200-15. doi: 10.1128/mBio.02200-15. mBio. 2016. PMID: 27073097 Free PMC article.
-
Widespread interspecies homologous recombination reveals reticulate evolution within the genus Streptomyces.Mol Phylogenet Evol. 2016 Sep;102:246-54. doi: 10.1016/j.ympev.2016.06.004. Epub 2016 Jun 18. Mol Phylogenet Evol. 2016. PMID: 27329941
-
Widespread homologous recombination within and between Streptomyces species.ISME J. 2010 Sep;4(9):1136-43. doi: 10.1038/ismej.2010.45. Epub 2010 Apr 15. ISME J. 2010. PMID: 20393569
-
Intraspecies comparison of Streptomyces pratensis genomes reveals high levels of recombination and gene conservation between strains of disparate geographic origin.BMC Genomics. 2014 Nov 15;15(1):970. doi: 10.1186/1471-2164-15-970. BMC Genomics. 2014. PMID: 25399205 Free PMC article.
-
Population Genomics Insights into Adaptive Evolution and Ecological Differentiation in Streptomycetes.Appl Environ Microbiol. 2019 Mar 22;85(7):e02555-18. doi: 10.1128/AEM.02555-18. Print 2019 Apr 1. Appl Environ Microbiol. 2019. PMID: 30658977 Free PMC article.
Cited by
-
Genetic Diversity and Anti-Oxidative Potential of Streptomyces spp. Isolated from Unexplored Niches of Meghalaya, India.Curr Microbiol. 2022 Nov 3;79(12):379. doi: 10.1007/s00284-022-03088-w. Curr Microbiol. 2022. PMID: 36329226
-
Within-Species Genomic Variation and Variable Patterns of Recombination in the Tetracycline Producer Streptomyces rimosus.Front Microbiol. 2019 Mar 21;10:552. doi: 10.3389/fmicb.2019.00552. eCollection 2019. Front Microbiol. 2019. PMID: 30949149 Free PMC article.
-
Exclusivity offers a sound yet practical species criterion for bacteria despite abundant gene flow.BMC Genomics. 2018 Oct 3;19(1):724. doi: 10.1186/s12864-018-5099-6. BMC Genomics. 2018. PMID: 30285620 Free PMC article.
-
Phylogenetic conservatism of thermal traits explains dispersal limitation and genomic differentiation of Streptomyces sister-taxa.ISME J. 2018 Sep;12(9):2176-2186. doi: 10.1038/s41396-018-0180-3. Epub 2018 Jun 7. ISME J. 2018. PMID: 29880909 Free PMC article.
-
Multidimensional insights into the biodiversity of Streptomyces in soils of China: a pilot study.Microbiol Spectr. 2025 Apr 2;13(5):e0169224. doi: 10.1128/spectrum.01692-24. Online ahead of print. Microbiol Spectr. 2025. PMID: 40172189 Free PMC article.
References
-
- Alacevic M. (1963). Interspecific recombination in Streptomyces. Nature 197: 1323. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. - PubMed
-
- Bapteste E, Boucher Y. (2009). Epistemological impacts of horizontal gene transfer on classification in microbiology. Methods Mol Biol Clifton NJ 532: 55–72. - PubMed
-
- Barilani M, Bernard-Laurent A, Mucci N, Tabarroni C, Kark S, Perez Garrido JA et al. (2007). Hybridisation with introduced chukars (Alectoris chukar threatens the gene pool integrity of native rock (A. graeca and red-legged (A. rufa partridge populations. Biol Cons 137: 57–69.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

