Ataxin-2 (Atxn2)-Knock-Out Mice Show Branched Chain Amino Acids and Fatty Acids Pathway Alterations
- PMID: 26850065
- PMCID: PMC4858951
- DOI: 10.1074/mcp.M115.056770
Ataxin-2 (Atxn2)-Knock-Out Mice Show Branched Chain Amino Acids and Fatty Acids Pathway Alterations
Abstract
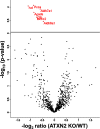
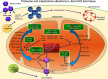
Human Ataxin-2 (ATXN2) gene locus variants have been associated with obesity, diabetes mellitus type 1,and hypertension in genome-wide association studies, whereas mouse studies showed the knock-out of Atxn2 to lead to obesity, insulin resistance, and dyslipidemia. Intriguingly, the deficiency of ATXN2 protein orthologs in yeast and flies rescues the neurodegeneration process triggered by TDP-43 and Ataxin-1 toxicity. To understand the molecular effects of ATXN2 deficiency by unbiased approaches, we quantified the global proteome and metabolome of Atxn2-knock-out mice with label-free mass spectrometry. In liver tissue, significant downregulations of the proteins ACADS, ALDH6A1, ALDH7A1, IVD, MCCC2, PCCA, OTC, together with bioinformatic enrichment of downregulated pathways for branched chain and other amino acid metabolism, fatty acids, and citric acid cycle were observed. Statistical trends in the cerebellar proteome and in the metabolomic profiles supported these findings. They are in good agreement with recent claims that PBP1, the yeast ortholog of ATXN2, sequestrates the nutrient sensor TORC1 in periods of cell stress. Overall, ATXN2 appears to modulate nutrition and metabolism, and its activity changes are determinants of growth excess or cell atrophy.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures


Similar articles
-
In Human and Mouse Spino-Cerebellar Tissue, Ataxin-2 Expansion Affects Ceramide-Sphingomyelin Metabolism.Int J Mol Sci. 2019 Nov 21;20(23):5854. doi: 10.3390/ijms20235854. Int J Mol Sci. 2019. PMID: 31766565 Free PMC article.
-
Atxn2 Knockout and CAG42-Knock-in Cerebellum Shows Similarly Dysregulated Expression in Calcium Homeostasis Pathway.Cerebellum. 2017 Feb;16(1):68-81. doi: 10.1007/s12311-016-0762-4. Cerebellum. 2017. PMID: 26868665 Free PMC article.
-
Calcium-responsive transactivator (CREST) toxicity is rescued by loss of PBP1/ATXN2 function in a novel yeast proteinopathy model and in transgenic flies.PLoS Genet. 2019 Aug 7;15(8):e1008308. doi: 10.1371/journal.pgen.1008308. eCollection 2019 Aug. PLoS Genet. 2019. PMID: 31390360 Free PMC article.
-
Efficient Prevention of Neurodegenerative Diseases by Depletion of Starvation Response Factor Ataxin-2.Trends Neurosci. 2017 Aug;40(8):507-516. doi: 10.1016/j.tins.2017.06.004. Epub 2017 Jul 3. Trends Neurosci. 2017. PMID: 28684172 Review.
-
Ataxin-2: A versatile posttranscriptional regulator and its implication in neural function.Wiley Interdiscip Rev RNA. 2018 Nov;9(6):e1488. doi: 10.1002/wrna.1488. Epub 2018 Jun 5. Wiley Interdiscip Rev RNA. 2018. PMID: 29869836 Review.
Cited by
-
In Human and Mouse Spino-Cerebellar Tissue, Ataxin-2 Expansion Affects Ceramide-Sphingomyelin Metabolism.Int J Mol Sci. 2019 Nov 21;20(23):5854. doi: 10.3390/ijms20235854. Int J Mol Sci. 2019. PMID: 31766565 Free PMC article.
-
Cell size and fat content of dietary-restricted Caenorhabditis elegans are regulated by ATX-2, an mTOR repressor.Proc Natl Acad Sci U S A. 2016 Aug 9;113(32):E4620-9. doi: 10.1073/pnas.1512156113. Epub 2016 Jul 25. Proc Natl Acad Sci U S A. 2016. PMID: 27457958 Free PMC article.
-
Unveiling the shared genetic architecture between testosterone and polycystic ovary syndrome.Sci Rep. 2024 Oct 13;14(1):23931. doi: 10.1038/s41598-024-75816-0. Sci Rep. 2024. PMID: 39397165 Free PMC article.
-
Additional common loci associated with stroke and obesity identified using pleiotropic analytical approach.Mol Genet Genomics. 2020 Mar;295(2):439-451. doi: 10.1007/s00438-019-01630-3. Epub 2019 Dec 7. Mol Genet Genomics. 2020. PMID: 31813042
-
Integrated view and comparative analysis of baseline protein expression in mouse and rat tissues.PLoS Comput Biol. 2022 Jun 17;18(6):e1010174. doi: 10.1371/journal.pcbi.1010174. eCollection 2022 Jun. PLoS Comput Biol. 2022. PMID: 35714157 Free PMC article.
References
-
- Huynh D. P., Del Bigio M. R., Ho D. H., and Pulst S. M. (1999) Expression of ataxin-2 in brains from normal individuals and patients with Alzheimer's disease and spinocerebellar ataxia 2. Ann. Neurol. 45, 232–241 - PubMed
-
- Fittschen M., Lastres-Becker I., Halbach M. V., Damrath E., Gispert S., Azizov M., Walter M., Muller S., and Auburger G. (2015) Genetic ablation of ataxin-2 increases several global translation factors in their transcript abundance but decreases translation rate. Neurogenetics 16, 181–192 - PMC - PubMed
-
- Sahba S., Nechiporuk A., Figueroa K. P., Nechiporuk T., and Pulst S. M. (1998) Genomic structure of the human gene for spinocerebellar ataxia type 2 (SCA2) on chromosome 12q24.1. Genomics 47, 359–364 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

